[English] 日本語
 Yorodumi
Yorodumi- EMDB-22080: EM Structure of Full-Length Androgen Receptor Coactivator Complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22080 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EM Structure of Full-Length Androgen Receptor Coactivator Complex | |||||||||
 Map data Map data | AR/ARE-DNA/p300/SRC-3 complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
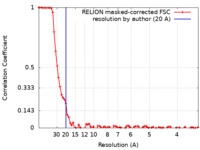
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Yu X / Yi P | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Structural Insights of Transcriptionally Active, Full-Length Androgen Receptor Coactivator Complexes. Authors: Xinzhe Yu / Ping Yi / Ross A Hamilton / Hong Shen / Muyuan Chen / Charles E Foulds / Michael A Mancini / Steven J Ludtke / Zhao Wang / Bert W O'Malley /  Abstract: Steroid receptors activate gene transcription by recruiting coactivators to initiate transcription of their target genes. For most nuclear receptors, the ligand-dependent activation function domain-2 ...Steroid receptors activate gene transcription by recruiting coactivators to initiate transcription of their target genes. For most nuclear receptors, the ligand-dependent activation function domain-2 (AF-2) is a primary contributor to the nuclear receptor (NR) transcriptional activity. In contrast to other steroid receptors, such as ERα, the activation function of androgen receptor (AR) is largely dependent on its ligand-independent AF-1 located in its N-terminal domain (NTD). It remains unclear why AR utilizes a different AF domain from other receptors despite that NRs share similar domain organizations. Here, we present cryoelectron microscopy (cryo-EM) structures of DNA-bound full-length AR and its complex structure with key coactivators, SRC-3 and p300. AR dimerization follows a unique head-to-head and tail-to-tail manner. Unlike ERα, AR directly contacts a single SRC-3 and p300. The AR NTD is the primary site for coactivator recruitment. The structures provide a basis for understanding assembly of the AR:coactivator complex and its domain contributions for coactivator assembly and transcriptional regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22080.map.gz emd_22080.map.gz | 37.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22080-v30.xml emd-22080-v30.xml emd-22080.xml emd-22080.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22080_fsc.xml emd_22080_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_22080.png emd_22080.png | 42.5 KB | ||
| Others |  emd_22080_additional_1.map.gz emd_22080_additional_1.map.gz emd_22080_additional_2.map.gz emd_22080_additional_2.map.gz | 23.3 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22080 http://ftp.pdbj.org/pub/emdb/structures/EMD-22080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22080 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22080 | HTTPS FTP |
-Validation report
| Summary document |  emd_22080_validation.pdf.gz emd_22080_validation.pdf.gz | 78.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22080_full_validation.pdf.gz emd_22080_full_validation.pdf.gz | 77.8 KB | Display | |
| Data in XML |  emd_22080_validation.xml.gz emd_22080_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22080 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22080 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22080 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22080 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10458 (Title: Structural Insights of Transcriptionally Active, Full-Length Androgen Receptor Coactivator Complexes EMPIAR-10458 (Title: Structural Insights of Transcriptionally Active, Full-Length Androgen Receptor Coactivator ComplexesData size: 5.7 TB / Data #1: ARE-DNA/AR/p300/SRC-3 [micrographs - multiframe] / Data #2: ARE-DNA/AR/p300/SRC-3 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22080.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22080.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AR/ARE-DNA/p300/SRC-3 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.74 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: AR/ARE-DNA/p300/SRC-3 binding with SRC3-Ab
| File | emd_22080_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AR/ARE-DNA/p300/SRC-3 binding with SRC3-Ab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: AR/ARE-DNA/p300/SRC-3 binding with AR-Ab1
| File | emd_22080_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AR/ARE-DNA/p300/SRC-3 binding with AR-Ab1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AR/ARE-DNA/p300/SRC-3 complex
| Entire | Name: AR/ARE-DNA/p300/SRC-3 complex |
|---|---|
| Components |
|
-Supramolecule #1: AR/ARE-DNA/p300/SRC-3 complex
| Supramolecule | Name: AR/ARE-DNA/p300/SRC-3 complex / type: complex / ID: 1 / Parent: 0 / Details: Androgen Receptor coactivator complex |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Molecular weight | Experimental: 800 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 5741 / Average exposure time: 10.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)