+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4243 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High-resolution cryo-EM structure of the human 80S ribosome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
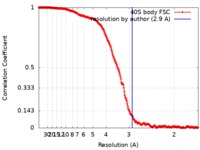
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Natchiar SK / Myasnikov AG | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Visualization of chemical modifications in the human 80S ribosome structure. Authors: S Kundhavai Natchiar / Alexander G Myasnikov / Hanna Kratzat / Isabelle Hazemann / Bruno P Klaholz /  Abstract: Chemical modifications of human ribosomal RNA (rRNA) are introduced during biogenesis and have been implicated in the dysregulation of protein synthesis, as is found in cancer and other diseases. ...Chemical modifications of human ribosomal RNA (rRNA) are introduced during biogenesis and have been implicated in the dysregulation of protein synthesis, as is found in cancer and other diseases. However, their role in this phenomenon is unknown. Here we visualize more than 130 individual rRNA modifications in the three-dimensional structure of the human ribosome, explaining their structural and functional roles. In addition to a small number of universally conserved sites, we identify many eukaryote- or human-specific modifications and unique sites that form an extended shell in comparison to bacterial ribosomes, and which stabilize the RNA. Several of the modifications are associated with the binding sites of three ribosome-targeting antibiotics, or are associated with degenerate states in cancer, such as keto alkylations on nucleotide bases reminiscent of specialized ribosomes. This high-resolution structure of the human 80S ribosome paves the way towards understanding the role of epigenetic rRNA modifications in human diseases and suggests new possibilities for designing selective inhibitors and therapeutic drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4243.map.gz emd_4243.map.gz | 61.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4243-v30.xml emd-4243-v30.xml emd-4243.xml emd-4243.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4243_fsc.xml emd_4243_fsc.xml | 22.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_4243.png emd_4243.png | 237.5 KB | ||
| Masks |  emd_4243_msk_1.map emd_4243_msk_1.map | 1000 MB |  Mask map Mask map | |
| Others |  emd_4243_additional.map.gz emd_4243_additional.map.gz emd_4243_half_map_1.map.gz emd_4243_half_map_1.map.gz emd_4243_half_map_2.map.gz emd_4243_half_map_2.map.gz | 811.2 MB 808.8 MB 810 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4243 http://ftp.pdbj.org/pub/emdb/structures/EMD-4243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4243 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4243.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4243.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4243_msk_1.map emd_4243_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_4243_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_4243_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_4243_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 80S ribosome
| Entire | Name: 80S ribosome |
|---|---|
| Components |
|
-Supramolecule #1: 80S ribosome
| Supramolecule | Name: 80S ribosome / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Experimental: 4.3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 100mM KCl, 5mM MgAc2, 20mM Hepes, 10mM NH4Cl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)