+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21688 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human TRPA1 in complex with inhibitor GDC-0334 | |||||||||
 Map data Map data | Main map, used for making figures. Sharpened, FOM-filtered, unmasked. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPA1 / channel / inhibitor / antagonist / MEMBRANE PROTEIN / MEMBRANE PROTEIN-Inhibitor complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtemperature-gated cation channel activity / stereocilium bundle / detection of chemical stimulus involved in sensory perception of pain / thermoception / TRP channels / response to pain / intracellularly gated calcium channel activity / detection of mechanical stimulus involved in sensory perception of pain / monoatomic ion transport / sensory perception of pain ...temperature-gated cation channel activity / stereocilium bundle / detection of chemical stimulus involved in sensory perception of pain / thermoception / TRP channels / response to pain / intracellularly gated calcium channel activity / detection of mechanical stimulus involved in sensory perception of pain / monoatomic ion transport / sensory perception of pain / response to cold / calcium ion transmembrane transport / calcium channel activity / response to organic cyclic compound / cellular response to hydrogen peroxide / intracellular calcium ion homeostasis / channel activity / protein homotetramerization / cell surface receptor signaling pathway / response to xenobiotic stimulus / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
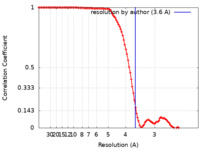
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Rohou A / Rouge L / Arthur CP | |||||||||
 Citation Citation |  Journal: J Exp Med / Year: 2021 Journal: J Exp Med / Year: 2021Title: A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment. Authors: Alessia Balestrini / Victory Joseph / Michelle Dourado / Rebecca M Reese / Shannon D Shields / Lionel Rougé / Daniel D Bravo / Tania Chernov-Rogan / Cary D Austin / Huifen Chen / Lan Wang / ...Authors: Alessia Balestrini / Victory Joseph / Michelle Dourado / Rebecca M Reese / Shannon D Shields / Lionel Rougé / Daniel D Bravo / Tania Chernov-Rogan / Cary D Austin / Huifen Chen / Lan Wang / Elisia Villemure / Daniel G M Shore / Vishal A Verma / Baihua Hu / Yong Chen / Laurie Leong / Chris Bjornson / Kathy Hötzel / Alvin Gogineni / Wyne P Lee / Eric Suto / Xiumin Wu / John Liu / Juan Zhang / Vineela Gandham / Jianyong Wang / Jian Payandeh / Claudio Ciferri / Alberto Estevez / Christopher P Arthur / Jens Kortmann / Ryan L Wong / Jose E Heredia / Jonas Doerr / Min Jung / Jason A Vander Heiden / Merone Roose-Girma / Lucinda Tam / Kai H Barck / Richard A D Carano / Han Ting Ding / Bobby Brillantes / Christine Tam / Xiaoying Yang / Simon S Gao / Justin Q Ly / Liling Liu / Liuxi Chen / Bianca M Liederer / Joseph H Lin / Steven Magnuson / Jun Chen / David H Hackos / Justin Elstrott / Alexis Rohou / Brian S Safina / Matthew Volgraf / Rebecca N Bauer / Lorena Riol-Blanco /  Abstract: Despite the development of effective therapies, a substantial proportion of asthmatics continue to have uncontrolled symptoms, airflow limitation, and exacerbations. Transient receptor potential ...Despite the development of effective therapies, a substantial proportion of asthmatics continue to have uncontrolled symptoms, airflow limitation, and exacerbations. Transient receptor potential cation channel member A1 (TRPA1) agonists are elevated in human asthmatic airways, and in rodents, TRPA1 is involved in the induction of airway inflammation and hyperreactivity. Here, the discovery and early clinical development of GDC-0334, a highly potent, selective, and orally bioavailable TRPA1 antagonist, is described. GDC-0334 inhibited TRPA1 function on airway smooth muscle and sensory neurons, decreasing edema, dermal blood flow (DBF), cough, and allergic airway inflammation in several preclinical species. In a healthy volunteer Phase 1 study, treatment with GDC-0334 reduced TRPA1 agonist-induced DBF, pain, and itch, demonstrating GDC-0334 target engagement in humans. These data provide therapeutic rationale for evaluating TRPA1 inhibition as a clinical therapy for asthma. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21688.map.gz emd_21688.map.gz | 95.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21688-v30.xml emd-21688-v30.xml emd-21688.xml emd-21688.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21688_fsc.xml emd_21688_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_21688.png emd_21688.png | 175.9 KB | ||
| Filedesc metadata |  emd-21688.cif.gz emd-21688.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21688 http://ftp.pdbj.org/pub/emdb/structures/EMD-21688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21688 | HTTPS FTP |
-Validation report
| Summary document |  emd_21688_validation.pdf.gz emd_21688_validation.pdf.gz | 659.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21688_full_validation.pdf.gz emd_21688_full_validation.pdf.gz | 658.9 KB | Display | |
| Data in XML |  emd_21688_validation.xml.gz emd_21688_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_21688_validation.cif.gz emd_21688_validation.cif.gz | 15.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21688 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21688 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21688 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21688 | HTTPS FTP |
-Related structure data
| Related structure data |  6wj5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21688.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21688.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map, used for making figures. Sharpened, FOM-filtered, unmasked. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TRPA1 bound by inhibitor GDC-0334
| Entire | Name: TRPA1 bound by inhibitor GDC-0334 |
|---|---|
| Components |
|
-Supramolecule #1: TRPA1 bound by inhibitor GDC-0334
| Supramolecule | Name: TRPA1 bound by inhibitor GDC-0334 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transient receptor potential cation channel subfamily A member 1
| Macromolecule | Name: Transient receptor potential cation channel subfamily A member 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.622125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPLHFAASYG RINTCQRLLQ DISDTRLLNE GDLHGMTPLH LAAKNGHDKV VQLLLKKGAL FLSDHNGWTA LHHASMGGYT QTMKVILDT NLKCTDRLDE DGNTALHFAA REGHAKAVAL LLSHNADIVL NKQQASFLHL ALHNKRKEVV LTIIRSKRWD E CLKIFSHN ...String: SPLHFAASYG RINTCQRLLQ DISDTRLLNE GDLHGMTPLH LAAKNGHDKV VQLLLKKGAL FLSDHNGWTA LHHASMGGYT QTMKVILDT NLKCTDRLDE DGNTALHFAA REGHAKAVAL LLSHNADIVL NKQQASFLHL ALHNKRKEVV LTIIRSKRWD E CLKIFSHN SPGNKCPITE MIEYLPECMK VLLDFCMLHS TEDKSCRDYY IEYNFKYLQC PLEFTKKTPT QDVIYEPLTA LN AMVQNNR IELLNHPVCK EYLLMKWLAY GFRAHMMNLG SYCLGLIPMT ILVVNIKPGM AFNSTGIINE TSDHSEILDT TNS YLIKTC MILVFLSSIF GYCKEAGQIF QQKRNYFMDI SNVLEWIIYT TGIIFVLPLF VEIPAHLQWQ CGAIAVYFYW MNFL LYLQR FENCGIFIVM LEVILKTLLR STVVFIFLLL AFGLSFYILL NLQDPFSSPL LSIIQTFSMM LGDINYRESF LEPYL RNEL AHPVLSFAQL VSFTIFVPIV LMNLLIGLAV GDIADVQKHA SLKRIAMQVE LHTSLEKKLP LWFLRKVDQK STIVYP NKP RSGGMLFHIF CFLFCTGEIR QEIPNADKSL EMEILKQKYR LKDLTFLLEK QHELIKLIIQ KMEIISET UniProtKB: Transient receptor potential cation channel subfamily A member 1 |
-Macromolecule #2: (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(tri...
| Macromolecule | Name: (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide type: ligand / ID: 2 / Number of copies: 4 / Formula: LXY |
|---|---|
| Molecular weight | Theoretical: 609.492 Da |
| Chemical component information | 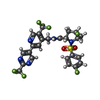 ChemComp-LXY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.33 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.2 Component:
| |||||||||||||||
| Grid | Model: C-flat-2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.067 kPa Details: Grid was gold-coated. Glow discharge using GloQube, negative polarity. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 2 x 4s blot time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 47.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | CCG MOE was used for refinement after Phenix real-space refinement. |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-6wj5: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)