+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20956 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Portal vertex structure of bacteriophage T4 | |||||||||
 Map data Map data | low-pass filtered and B-factor sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | portal protein assembly / gp20 / gp23 / portal vertex / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont genome ejection through host cell envelope, contractile tail mechanism / T=13 icosahedral viral capsid / viral portal complex / viral genome packaging / viral release from host cell / viral capsid / host cell plasma membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Enterobacteria phage T4 (virus) / Enterobacteria phage T4 (virus) /  Escherichia virus T4 Escherichia virus T4 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Fang Q / Fokine A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structural morphing in a symmetry-mismatched viral vertex. Authors: Qianglin Fang / Wei-Chun Tang / Pan Tao / Marthandan Mahalingam / Andrei Fokine / Michael G Rossmann / Venigalla B Rao /  Abstract: Large biological structures are assembled from smaller, often symmetric, sub-structures. However, asymmetry among sub-structures is fundamentally important for biological function. An extreme form of ...Large biological structures are assembled from smaller, often symmetric, sub-structures. However, asymmetry among sub-structures is fundamentally important for biological function. An extreme form of asymmetry, a 12-fold-symmetric dodecameric portal complex inserted into a 5-fold-symmetric capsid vertex, is found in numerous icosahedral viruses, including tailed bacteriophages, herpesviruses, and archaeal viruses. This vertex is critical for driving capsid assembly, DNA packaging, tail attachment, and genome ejection. Here, we report the near-atomic in situ structure of the symmetry-mismatched portal vertex from bacteriophage T4. Remarkably, the local structure of portal morphs to compensate for symmetry-mismatch, forming similar interactions in different capsid environments while maintaining strict symmetry in the rest of the structure. This creates a unique and unusually dynamic symmetry-mismatched vertex that is central to building an infectious virion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20956.map.gz emd_20956.map.gz | 16.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20956-v30.xml emd-20956-v30.xml emd-20956.xml emd-20956.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20956.png emd_20956.png | 275.6 KB | ||
| Filedesc metadata |  emd-20956.cif.gz emd-20956.cif.gz | 5.5 KB | ||
| Others |  emd_20956_additional_1.map.gz emd_20956_additional_1.map.gz emd_20956_additional_2.map.gz emd_20956_additional_2.map.gz | 24.3 MB 916.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20956 http://ftp.pdbj.org/pub/emdb/structures/EMD-20956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20956 | HTTPS FTP |
-Validation report
| Summary document |  emd_20956_validation.pdf.gz emd_20956_validation.pdf.gz | 393.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20956_full_validation.pdf.gz emd_20956_full_validation.pdf.gz | 393 KB | Display | |
| Data in XML |  emd_20956_validation.xml.gz emd_20956_validation.xml.gz | 7.9 KB | Display | |
| Data in CIF |  emd_20956_validation.cif.gz emd_20956_validation.cif.gz | 9.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20956 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20956 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20956 | HTTPS FTP |
-Related structure data
| Related structure data |  6uzcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20956.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20956.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | low-pass filtered and B-factor sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.44 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
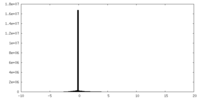
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
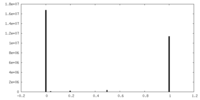
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unfiltered map
| File | emd_20956_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
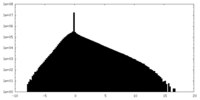
| Density Histograms |
-Supplemental map: emd 20956 additional 2.map
| File | emd_20956_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
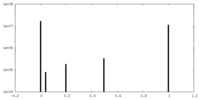
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia virus T4
| Entire | Name:  Escherichia virus T4 Escherichia virus T4 |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia virus T4
| Supramolecule | Name: Escherichia virus T4 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10665 / Sci species name: Escherichia virus T4 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 30 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
| Molecular weight | Theoretical: 56.074242 KDa |
| Sequence | String: MTIKTKAELL NKWKPLLEGE GLPEIANSKQ AIIAKIFENQ EKDFQTAPEY KDEKIAQAFG SFLTEAEIGG DHGYNATNIA AGQTSGAVT QIGPAVMGMV RRAIPNLIAF DICGVQPMNS PTGQVFALRA VYGKDPVAAG AKEAFHPMYG PDAMFSGQGA A KKFPALAA ...String: MTIKTKAELL NKWKPLLEGE GLPEIANSKQ AIIAKIFENQ EKDFQTAPEY KDEKIAQAFG SFLTEAEIGG DHGYNATNIA AGQTSGAVT QIGPAVMGMV RRAIPNLIAF DICGVQPMNS PTGQVFALRA VYGKDPVAAG AKEAFHPMYG PDAMFSGQGA A KKFPALAA STQTTVGDIY THFFQETGTV YLQASVQVTI DAGATDAAKL DAEIKKQMEA GALVEIAEGM ATSIAELQEG FN GSTDNPW NEMGFRIDKQ VIEAKSRQLK AAYSIELAQD LRAVHGMDAD AELSGILATE IMLEINREVV DWINYSAQVG KSG MTLTPG SKAGVFDFQD PIDIRGARWA GESFKALLFQ IDKEAVEIAR QTGRGEGNFI IASRNVVNVL ASVDTGISYA AQGL ATGFS TDTTKSVFAG VLGGKYRVYI DQYAKQDYFT VGYKGPNEMD AGIYYAPYVA LTPLRGSDPK NFQPVMGFKT RYGIG INPF AESAAQAPAS RIQSGMPSIL NSLGKNAYFR RVYVKGI UniProtKB: Major capsid protein |
-Macromolecule #2: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
| Molecular weight | Theoretical: 61.117082 KDa |
| Sequence | String: MKFNVLSLFA PWAKMDERNF KDQEKEDLVS ITAPKLDDGA REFEVSSNEA ASPYNAAFQT IFGSYEPGMK TTRELIDTYR NLMNNYEVD NAVSEIVSDA IVYEDDTEVV ALNLDKSKFS PKIKNMMLDE FSDVLNHLSF QRKGSDHFRR WYVDSRIFFH K IIDPKRPK ...String: MKFNVLSLFA PWAKMDERNF KDQEKEDLVS ITAPKLDDGA REFEVSSNEA ASPYNAAFQT IFGSYEPGMK TTRELIDTYR NLMNNYEVD NAVSEIVSDA IVYEDDTEVV ALNLDKSKFS PKIKNMMLDE FSDVLNHLSF QRKGSDHFRR WYVDSRIFFH K IIDPKRPK EGIKELRRLD PRQVQYVREI ITETEAGTKI VKGYKEYFIY DTAHESYACD GRMYEAGTKI KIPKAAVVYA HS GLVDCCG KNIIGYLHRA VKPANQLKLL EDAVVIYRIT RAPDRRVWYV DTGNMPARKA AEHMQHVMNT MKNRVVYDAS TGK IKNQQH NMSMTEDYWL QRRDGKAVTE VDTLPGADNT GNMEDIRWFR QALYMALRVP LSRIPQDQQG GVMFDSGTSI TRDE LTFAK FIRELQHKFE EVFLDPLKTN LLLKGIITED EWNDEINNIK IEFHRDSYFA ELKEAEILER RINMLTMAEP FIGKY ISHR TAMKDILQMT DEEIEQEAKQ IEEESKEARF QDPDQEQEDF UniProtKB: Portal protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 23.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 53608 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6uzc: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)