+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20853 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | P7A7 ribosome large subunit | ||||||||||||
 Map data Map data | RELION post-processed map | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | ||||||||||||
 Authors Authors | Watson ZL / Ward FR / Cate JHD | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2019 Journal: Biochemistry / Year: 2019Title: Defects in the Assembly of Ribosomes Selected for β-Amino Acid Incorporation. Authors: Fred R Ward / Zoe L Watson / Omer Ad / Alanna Schepartz / Jamie H D Cate /  Abstract: Ribosome engineering has emerged as a promising field in synthetic biology, particularly concerning the production of new sequence-defined polymers. Mutant ribosomes have been developed that improve ...Ribosome engineering has emerged as a promising field in synthetic biology, particularly concerning the production of new sequence-defined polymers. Mutant ribosomes have been developed that improve the incorporation of several nonstandard monomers including d-amino acids, dipeptides, and β-amino acids into polypeptide chains. However, there remains little mechanistic understanding of how these ribosomes catalyze incorporation of these new substrates. Here, we probed the properties of a mutant ribosome-P7A7-evolved for better β-amino acid incorporation through biochemistry and cryo-electron microscopy. Although P7A7 is a functional ribosome , it is inactive , and assembles poorly into 70S ribosome complexes. Structural characterization revealed large regions of disorder in the peptidyltransferase center and nearby features, suggesting a defect in assembly. Comparison of RNA helix and ribosomal protein occupancy with other assembly intermediates revealed that P7A7 is stalled at a late stage in ribosome assembly, explaining its weak activity. These results highlight the importance of ensuring efficient ribosome assembly during ribosome engineering toward new catalytic abilities. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20853.map.gz emd_20853.map.gz | 92.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20853-v30.xml emd-20853-v30.xml emd-20853.xml emd-20853.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20853_fsc.xml emd_20853_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_20853.png emd_20853.png | 74 KB | ||
| Others |  emd_20853_additional.map.gz emd_20853_additional.map.gz emd_20853_additional_1.map.gz emd_20853_additional_1.map.gz emd_20853_half_map_1.map.gz emd_20853_half_map_1.map.gz emd_20853_half_map_2.map.gz emd_20853_half_map_2.map.gz | 77.3 MB 77.3 MB 77.5 MB 77.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20853 http://ftp.pdbj.org/pub/emdb/structures/EMD-20853 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20853 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20853 | HTTPS FTP |
-Validation report
| Summary document |  emd_20853_validation.pdf.gz emd_20853_validation.pdf.gz | 78.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20853_full_validation.pdf.gz emd_20853_full_validation.pdf.gz | 77.4 KB | Display | |
| Data in XML |  emd_20853_validation.xml.gz emd_20853_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20853 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20853 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20853 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20853 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20853.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20853.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1136 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
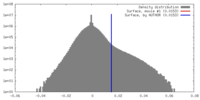
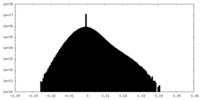
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: RELION 3d-autorefine map
| File | emd_20853_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION 3d-autorefine map | ||||||||||||
| Projections & Slices |
| ||||||||||||
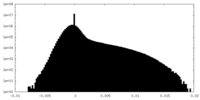
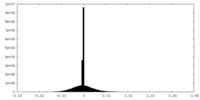
| Density Histograms |
-Additional map: RELION 3d-autorefine map
| File | emd_20853_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION 3d-autorefine map | ||||||||||||
| Projections & Slices |
| ||||||||||||
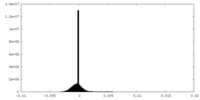
| Density Histograms |
-Half map: Half-map 1
| File | emd_20853_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
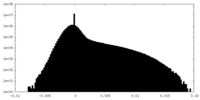
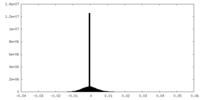
| Density Histograms |
-Half map: Half-map 2
| File | emd_20853_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : P7A7 E. coli 50S mutant
| Entire | Name: P7A7 E. coli 50S mutant |
|---|---|
| Components |
|
-Supramolecule #1: P7A7 E. coli 50S mutant
| Supramolecule | Name: P7A7 E. coli 50S mutant / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 10.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)