[English] 日本語
 Yorodumi
Yorodumi- EMDB-20240: Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20240 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | |||||||||
 Map data Map data | O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / TRANSFERASE / glycosylation | |||||||||
| Function / homology |  Function and homology information Function and homology informationdolichyl-phosphate-mannose-protein mannosyltransferase Pmt1p-Pmt3p dimer complex / dolichyl-phosphate-mannose-protein mannosyltransferase Pmt1p-Pmt2p dimer complex / dolichyl-phosphate-mannose-protein mannosyltransferase Pmt5p-Pmt2p dimer complex / dolichyl-phosphate-mannose-protein mannosyltransferase / dolichyl-phosphate-mannose-protein mannosyltransferase activity / regulation of endoplasmic reticulum unfolded protein response / fungal-type cell wall biogenesis / protein O-linked mannosylation / protein exit from endoplasmic reticulum / protein O-linked glycosylation ...dolichyl-phosphate-mannose-protein mannosyltransferase Pmt1p-Pmt3p dimer complex / dolichyl-phosphate-mannose-protein mannosyltransferase Pmt1p-Pmt2p dimer complex / dolichyl-phosphate-mannose-protein mannosyltransferase Pmt5p-Pmt2p dimer complex / dolichyl-phosphate-mannose-protein mannosyltransferase / dolichyl-phosphate-mannose-protein mannosyltransferase activity / regulation of endoplasmic reticulum unfolded protein response / fungal-type cell wall biogenesis / protein O-linked mannosylation / protein exit from endoplasmic reticulum / protein O-linked glycosylation / ERAD pathway / endoplasmic reticulum membrane / endoplasmic reticulum Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Bai L / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex. Authors: Lin Bai / Amanda Kovach / Qinglong You / Alanna Kenny / Huilin Li /  Abstract: In eukaryotes, a nascent peptide entering the endoplasmic reticulum (ER) is scanned by two Sec61 translocon-associated large membrane machines for protein N-glycosylation and protein O-mannosylation, ...In eukaryotes, a nascent peptide entering the endoplasmic reticulum (ER) is scanned by two Sec61 translocon-associated large membrane machines for protein N-glycosylation and protein O-mannosylation, respectively. While the structure of the eight-protein oligosaccharyltransferase complex has been determined recently, the structures of mannosyltransferases of the PMT family, which are an integral part of ER protein homeostasis, are still unknown. Here we report cryo-EM structures of the Saccharomyces cerevisiae Pmt1-Pmt2 complex bound to a donor and an acceptor peptide at 3.2-Å resolution, showing that each subunit contains 11 transmembrane helices and a lumenal β-trefoil fold termed the MIR domain. The structures reveal the substrate recognition model and confirm an inverting mannosyl-transferring reaction mechanism by the enzyme complex. Furthermore, we found that the transmembrane domains of Pmt1 and Pmt2 share a structural fold with the catalytic subunits of oligosaccharyltransferases, confirming a previously proposed evolutionary relationship between protein O-mannosylation and protein N-glycosylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20240.map.gz emd_20240.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20240-v30.xml emd-20240-v30.xml emd-20240.xml emd-20240.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20240.png emd_20240.png | 241.3 KB | ||
| Filedesc metadata |  emd-20240.cif.gz emd-20240.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20240 http://ftp.pdbj.org/pub/emdb/structures/EMD-20240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20240 | HTTPS FTP |
-Validation report
| Summary document |  emd_20240_validation.pdf.gz emd_20240_validation.pdf.gz | 389.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20240_full_validation.pdf.gz emd_20240_full_validation.pdf.gz | 389.4 KB | Display | |
| Data in XML |  emd_20240_validation.xml.gz emd_20240_validation.xml.gz | 5.8 KB | Display | |
| Data in CIF |  emd_20240_validation.cif.gz emd_20240_validation.cif.gz | 6.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20240 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20240 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20240 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20240 | HTTPS FTP |
-Related structure data
| Related structure data |  6p2rMC  6p25C  6p28C  20237 C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20240.map.gz / Format: CCP4 / Size: 37.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20240.map.gz / Format: CCP4 / Size: 37.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.029 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
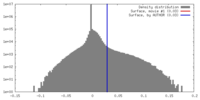
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Protein O-mannosyl transferase complex
| Entire | Name: Protein O-mannosyl transferase complex |
|---|---|
| Components |
|
-Supramolecule #1: Protein O-mannosyl transferase complex
| Supramolecule | Name: Protein O-mannosyl transferase complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Dolichyl-phosphate-mannose--protein mannosyltransferase 1
| Macromolecule | Name: Dolichyl-phosphate-mannose--protein mannosyltransferase 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: dolichyl-phosphate-mannose-protein mannosyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.771125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSEEKTYKRV EQDDPVPELD IKQGPVRPFI VTDPSAELAS LRTMVTLKEK LLVACLAVFT AVIRLHGLAW PDSVVFDEVH FGGFASQYI RGTYFMDVHP PLAKMLYAGV ASLGGFQGDF DFENIGDSFP STTPYVLMRF FSASLGALTV ILMYMTLRYS G VRMWVALM ...String: MSEEKTYKRV EQDDPVPELD IKQGPVRPFI VTDPSAELAS LRTMVTLKEK LLVACLAVFT AVIRLHGLAW PDSVVFDEVH FGGFASQYI RGTYFMDVHP PLAKMLYAGV ASLGGFQGDF DFENIGDSFP STTPYVLMRF FSASLGALTV ILMYMTLRYS G VRMWVALM SAICFAVENS YVTISRYILL DAPLMFFIAA AVYSFKKYEM YPANSLNAYK SLLATGIALG MASSSKWVGL FT VTWVGLL CIWRLWFMIG DLTKSSKSIF KVAFAKLAFL LGVPFALYLV FFYIHFQSLT LDGDGASFFS PEFRSTLKNN KIP QNVVAD VGIGSIISLR HLSTMGGYLH SHSHNYPAGS EQQQSTLYPH MDANNDWLLE LYNAPGESLT TFQNLTDGTK VRLF HTVTR CRLHSHDHKP PVSESSDWQK EVSCYGYSGF DGDANDDWVV EIDKKNSAPG VAQERVIALD TKFRLRHAMT GCYLF SHEV KLPAWGFEQQ EVTCASSGRH DLTLWYVENN SNPLLPEDTK RISYKPASFI SKFIESHKKM WHINKNLVEP HVYESQ PTS WPFLLRGISY WGENNRNVYL LGNAIVWWAV TAFIGIFGLI VITELFSWQL GKPILKDSKV VNFHVQVIHY LLGFAVH YA PSFLMQRQMF LHHYLPAYYF GILALGHALD IIVSYVFRSK RQMGYAVVIT FLAASVYFFK SFSPIIYGTP WTQELCQK S QWLSGWDYNC NTYFSSLEEY KNQTLTKRES QPAATSTVEE ITIEGDGPSY EDLMNEDGKK IFKDTEGNEL DPEVVKKML EEEGANILKV EKRAVLE UniProtKB: Dolichyl-phosphate-mannose--protein mannosyltransferase 1 |
-Macromolecule #2: Dolichyl-phosphate-mannose--protein mannosyltransferase 2
| Macromolecule | Name: Dolichyl-phosphate-mannose--protein mannosyltransferase 2 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: dolichyl-phosphate-mannose-protein mannosyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.957422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSSSSTGYS KNNAAHIKQE NTLRQRESSS ISVSEELSSA DERDAEDFSK EKPAAQSSLL RLESVVMPVI FTALALFTRM YKIGINNHV VWDEAHFGKF GSYYLRHEFY HDVHPPLGKM LVGLSGYLAG YNGSWDFPSG EIYPDYLDYV KMRLFNASFS A LCVPLAYF ...String: MSSSSSTGYS KNNAAHIKQE NTLRQRESSS ISVSEELSSA DERDAEDFSK EKPAAQSSLL RLESVVMPVI FTALALFTRM YKIGINNHV VWDEAHFGKF GSYYLRHEFY HDVHPPLGKM LVGLSGYLAG YNGSWDFPSG EIYPDYLDYV KMRLFNASFS A LCVPLAYF TAKAIGFSLP TVWLMTVLVL FENSYSTLGR FILLDSMLLF FTVASFFSFV MFHNQRSKPF SRKWWKWLLI TG ISLGCTI SVKMVGLFII TMVGIYTVID LWTFLADKSM SWKTYINHWL ARIFGLIIVP FCIFLLCFKI HFDLLSHSGT GDA NMPSLF QARLVGSDVG QGPRDIALGS SVVSIKNQAL GGSLLHSHIQ TYPDGSNQQQ VTCYGYKDAN NEWFFNRERG LPSW SENET DIEYLKPGTS YRLVHKSTGR NLHTHPVAAP VSKTQWEVSG YGDNVVGDNK DNWVIEIMDQ RGDEDPEKLH TLTTS FRIK NLEMGCYLAQ TGNSLPEWGF RQQEVVCMKN PFKRDKRTWW NIETHENERL PPRPEDFQYP KTNFLKDFIH LNLAMM ATN NALVPDPDKF DYLASSAWQW PTLNVGLRLC GWGDDNPKYF LLGTPASTWA SSVAVLAFMA TVVILLIRWQ RQYVDLR NP SNWNVFLMGG FYPLLAWGLH YMPFVIMSRV TYVHHYLPAL YFALIILAYC FDAGLQKWSR SKCGRIMRFV LYAGFMAL V IGCFWYFSPI SFGMEGPSSN FRYLNWFSTW DIADKQEA UniProtKB: Dolichyl-phosphate-mannose--protein mannosyltransferase 2 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 4 / Number of copies: 3 / Formula: CPL |
|---|---|
| Molecular weight | Theoretical: 758.06 Da |
| Chemical component information |  ChemComp-CPL: |
-Macromolecule #5: (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-...
| Macromolecule | Name: (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate type: ligand / ID: 5 / Number of copies: 1 / Formula: NNM |
|---|---|
| Molecular weight | Theoretical: 644.947 Da |
| Chemical component information | 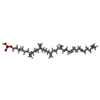 ChemComp-NNM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6p2r: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)