[English] 日本語
 Yorodumi
Yorodumi- EMDB-20205: Asymmetric focused reconstruction of human norovirus GI.1 Norwalk... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20205 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | |||||||||
 Map data Map data | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Caliciviridae / Norovirus / GI.1 / Norwalk / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=3 icosahedral viral capsid / host cell cytoplasm / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Norwalk virus (strain GI/Human/United States/Norwalk/1968) / Norwalk virus (strain GI/Human/United States/Norwalk/1968) /  Norovirus Hu/1968/US Norovirus Hu/1968/US | |||||||||
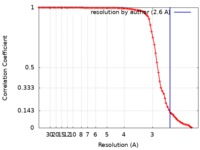
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Jung J / Grant T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations. Authors: James Jung / Timothy Grant / Dennis R Thomas / Chris W Diehnelt / Nikolaus Grigorieff / Leemor Joshua-Tor /  Abstract: Noroviruses are a leading cause of foodborne illnesses worldwide. Although GII.4 strains have been responsible for most norovirus outbreaks, the assembled virus shell structures have been available ...Noroviruses are a leading cause of foodborne illnesses worldwide. Although GII.4 strains have been responsible for most norovirus outbreaks, the assembled virus shell structures have been available in detail for only a single strain (GI.1). We present high-resolution (2.6- to 4.1-Å) cryoelectron microscopy (cryo-EM) structures of GII.4, GII.2, GI.7, and GI.1 human norovirus outbreak strain virus-like particles (VLPs). Although norovirus VLPs have been thought to exist in a single-sized assembly, our structures reveal polymorphism between and within genogroups, with small, medium, and large particle sizes observed. Using asymmetric reconstruction, we were able to resolve a Zn metal ion adjacent to the coreceptor binding site, which affected the structural stability of the shell. Our structures serve as valuable templates for facilitating vaccine formulations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20205.map.gz emd_20205.map.gz | 1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20205-v30.xml emd-20205-v30.xml emd-20205.xml emd-20205.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20205_fsc.xml emd_20205_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_20205.png emd_20205.png | 154.4 KB | ||
| Filedesc metadata |  emd-20205.cif.gz emd-20205.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20205 http://ftp.pdbj.org/pub/emdb/structures/EMD-20205 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20205 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20205 | HTTPS FTP |
-Validation report
| Summary document |  emd_20205_validation.pdf.gz emd_20205_validation.pdf.gz | 503.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20205_full_validation.pdf.gz emd_20205_full_validation.pdf.gz | 503 KB | Display | |
| Data in XML |  emd_20205_validation.xml.gz emd_20205_validation.xml.gz | 8.7 KB | Display | |
| Data in CIF |  emd_20205_validation.cif.gz emd_20205_validation.cif.gz | 11.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20205 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20205 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20205 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20205 | HTTPS FTP |
-Related structure data
| Related structure data |  6outMC  6otfC  6ou9C  6oucC  6ouuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20205.map.gz / Format: CCP4 / Size: 4.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20205.map.gz / Format: CCP4 / Size: 4.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Norovirus Hu/1968/US
| Entire | Name:  Norovirus Hu/1968/US Norovirus Hu/1968/US |
|---|---|
| Components |
|
-Supramolecule #1: Norovirus Hu/1968/US
| Supramolecule | Name: Norovirus Hu/1968/US / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 524364 / Sci species name: Norovirus Hu/1968/US / Sci species strain: GI.1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.19 MDa |
| Virus shell | Shell ID: 1 / Name: VP1 / Diameter: 410.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Norwalk virus (strain GI/Human/United States/Norwalk/1968) Norwalk virus (strain GI/Human/United States/Norwalk/1968)Strain: GI/Human/United States/Norwalk/1968 |
| Molecular weight | Theoretical: 56.629828 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMMASKDATS SVDGASGAGQ LVPEVNASDP LAMDPVAGSS TAVATAGQVN PIDPWIINNF VQAPQGEFTI SPNNTPGDVL FDLSLGPHL NPFLLHLSQM YNGWVGNMRV RIMLAGNAFT AGKIIVSCIP PGFGSHNLTI AQATLFPHVI ADVRTLDPIE V PLEDVRNV ...String: MMMASKDATS SVDGASGAGQ LVPEVNASDP LAMDPVAGSS TAVATAGQVN PIDPWIINNF VQAPQGEFTI SPNNTPGDVL FDLSLGPHL NPFLLHLSQM YNGWVGNMRV RIMLAGNAFT AGKIIVSCIP PGFGSHNLTI AQATLFPHVI ADVRTLDPIE V PLEDVRNV LFHNNDRNQQ TMRLVCMLYT PLRTGGGTGD SFVVAGRVMT CPSPDFNFLF LVPPTVEQKT RPFTLPNLPL SS LSNSRAP LPISSMGISP DNVQSVQFQN GRCTLDGRLV GTTPVSLSHV AKIRGTSNGT VINLTELDGT PFHPFEGPAP IGF PDLGGC DWHINMTQFG HSSQTQYDVD TTPDTFVPHL GSIQANGIGS GNYVGVLSWI SPPSHPSGSQ VDLWKIPNYG SSIT EATHL APSVYPPGFG EVLVFFMSKM PGPGAYNLPC LLPQEYISHL ASEQAPTVGE AALLHYVDPD TGRNLGEFKA YPDGF LTCV PNGASSGPQQ LPINGVFVFV SWVSRFYQLK PVGTASSARG RLGLRR UniProtKB: Capsid protein VP1 |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 226 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 5.75 |
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 1820 / Average exposure time: 7.0 sec. / Average electron dose: 78.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL | ||||||||
| Output model |  PDB-6out: |
 Movie
Movie Controller
Controller