[English] 日本語
 Yorodumi
Yorodumi- EMDB-20104: CDTb Double Heptamer Long Form Mask 1 Modeled from Cryo-EM Map Re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20104 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CDTb Double Heptamer Long Form Mask 1 Modeled from Cryo-EM Map Reconstructed using C7 Symmetry | |||||||||
 Map data Map data | CDTb Long Form Mask 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CDTb / Clostridium / Toxin / Binary / difficil / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein homooligomerization / transferase activity / extracellular region / identical protein binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) | |||||||||
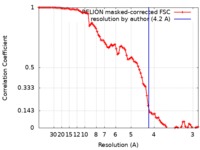
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Lacy DB / Sheedlo MJ | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2020 Journal: Nat Microbiol / Year: 2020Title: Structural insights into the transition of Clostridioides difficile binary toxin from prepore to pore. Authors: David M Anderson / Michael J Sheedlo / Jaime L Jensen / D Borden Lacy /  Abstract: Clostridioides (formerly Clostridium) difficile is a Gram-positive, spore-forming anaerobe and a leading cause of hospital-acquired infection and gastroenteritis-associated death in US hospitals. The ...Clostridioides (formerly Clostridium) difficile is a Gram-positive, spore-forming anaerobe and a leading cause of hospital-acquired infection and gastroenteritis-associated death in US hospitals. The disease state is usually preceded by disruption of the host microbiome in response to antibiotic treatment and is characterized by mild to severe diarrhoea. C. difficile infection is dependent on the secretion of one or more AB-type toxins: toxin A (TcdA), toxin B (TcdB) and the C. difficile transferase toxin (CDT). Whereas TcdA and TcdB are considered the primary virulence factors, recent studies suggest that CDT increases the severity of C. difficile infection in some of the most problematic clinical strains. To better understand how CDT functions, we used cryo-electron microscopy to define the structure of CDTb, the cell-binding component of CDT. We obtained structures of several oligomeric forms that highlight the conformational changes that enable conversion from a prepore to a β-barrel pore. The structural analysis also reveals a glycan-binding domain and residues involved in binding the host-cell receptor, lipolysis-stimulated lipoprotein receptor. Together, these results provide a framework to understand how CDT functions at the host cell interface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20104.map.gz emd_20104.map.gz | 55.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20104-v30.xml emd-20104-v30.xml emd-20104.xml emd-20104.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20104_fsc.xml emd_20104_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20104.png emd_20104.png | 41.8 KB | ||
| Filedesc metadata |  emd-20104.cif.gz emd-20104.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20104 http://ftp.pdbj.org/pub/emdb/structures/EMD-20104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20104 | HTTPS FTP |
-Validation report
| Summary document |  emd_20104_validation.pdf.gz emd_20104_validation.pdf.gz | 591.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20104_full_validation.pdf.gz emd_20104_full_validation.pdf.gz | 591 KB | Display | |
| Data in XML |  emd_20104_validation.xml.gz emd_20104_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_20104_validation.cif.gz emd_20104_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20104 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20104 | HTTPS FTP |
-Related structure data
| Related structure data |  6oktMC  0608C  0609C  0610C  6o2mC  6o2nC  6o2oC  6okrC  6oksC  6okuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20104.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20104.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CDTb Long Form Mask 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CDTb
| Entire | Name: CDTb |
|---|---|
| Components |
|
-Supramolecule #1: CDTb
| Supramolecule | Name: CDTb / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
-Macromolecule #1: ADP-ribosyltransferase binding component
| Macromolecule | Name: ADP-ribosyltransferase binding component / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) |
| Molecular weight | Theoretical: 98.916828 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIQMRNKKV LSFLTLTAIV SQALVYPVYA QTSTSNHSNK KKEIVNEDIL PNNGLMGYYF TDEHFKDLKL MAPIKDGNLK FEEKKVDKL LDKDKSDVKS IRWTGRIIPS KDGEYTLSTD RDDVLMQVNT ESTISNTLKV NMKKGKEYKV RIELQDKNLG S IDNLSSPN ...String: MKIQMRNKKV LSFLTLTAIV SQALVYPVYA QTSTSNHSNK KKEIVNEDIL PNNGLMGYYF TDEHFKDLKL MAPIKDGNLK FEEKKVDKL LDKDKSDVKS IRWTGRIIPS KDGEYTLSTD RDDVLMQVNT ESTISNTLKV NMKKGKEYKV RIELQDKNLG S IDNLSSPN LYWELDGMKK IIPEENLFLR DYSNIEKDDP FIPNNNFFDP KLMSDWEDED LDTDNDNIPD SYERNGYTIK DL IAVKWED SFAEQGYKKY VSNYLESNTA GDPYTDYEKA SGSFDKAIKT EARDPLVAAY PIVGVGMEKL IISTNEHAST DQG KTVSRA TTNSKTESNT AGVSVNVGYQ NGFTANVTTN YSHTTDNSTA VQDSNGESWN TGLSINKGES AYINANVRYY NTGT APMYK VTPTTNLVLD GDTLSTIKAQ ENQIGNNLSP GDTYPKKGLS PLALNTMDQF SSRLIPINYD QLKKLDAGKQ IKLET TQVS GNFGTKNSSG QIVTEGNSWS DYISQIDSIS ASIILDTENE SYERRVTAKN LQDPEDKTPE LTIGEAIEKA FGATKK DGL LYFNDIPIDE SCVELIFDDN TANKIKDSLK TLSDKKIYNV KLERGMNILI KTPTYFTNFD DYNNYPSTWS NVNTTNQ DG LQGSANKLNG ETKIKIPMSE LKPYKRYVFS GYSKDPLTSN SIIVKIKAKE EKTDYLVPEQ GYTKFSYEFE TTEKDSSN I EITLIGSGTT YLDNLSITEL NSTPEILDEP EVKIPTDQEI MDAHKIYFAD LNFNPSTGNT YINGMYFAPT QTNKEALDY IQKYRVEATL QYSGFKDIGT KDKEMRNYLG DPNQPKTNYV NLRSYFTGGE NIMTYKKLRI YAITPDDREL LVLSVD UniProtKB: ADP-ribosyltransferase binding component |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 2 / Number real images: 4914 / Average exposure time: 9.6 sec. / Average electron dose: 110.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 75 |
|---|---|
| Output model |  PDB-6okt: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)