[English] 日本語
 Yorodumi
Yorodumi- EMDB-19599: Structural characterization of Thogoto Virus nucleoprotein provid... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural characterization of Thogoto Virus nucleoprotein provides insights into RNA encapsidation and assembly | |||||||||
 Map data Map data | filtered to 18A, non-sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viral replication / nucleoprotein / RNA binding / oligomerization / orthomyxovirus / VIRAL PROTEIN | |||||||||
| Function / homology | helical viral capsid / viral penetration into host nucleus / host cell / viral nucleocapsid / ribonucleoprotein complex / symbiont entry into host cell / host cell nucleus / RNA binding / Nucleoprotein Function and homology information Function and homology information | |||||||||
| Biological species |  Thogotovirus Thogotovirus | |||||||||
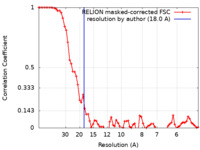
| Method | subtomogram averaging / cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Roske Y / Mikirtumov V / Daumke O / Kudryashev M / Dick A | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Structural characterization of Thogoto Virus nucleoprotein provides insights into viral RNA encapsidation and RNP assembly. Authors: Alexej Dick / Vasilii Mikirtumov / Jonas Fuchs / Ferdinand Krupp / Daniel Olal / Elias Bendl / Thiemo Sprink / Christoph Diebolder / Mikhail Kudryashev / Georg Kochs / Yvette Roske / Oliver Daumke /  Abstract: Orthomyxoviruses, such as influenza and thogotoviruses, are important human and animal pathogens. Their segmented viral RNA genomes are wrapped by viral nucleoproteins (NPs) into helical ...Orthomyxoviruses, such as influenza and thogotoviruses, are important human and animal pathogens. Their segmented viral RNA genomes are wrapped by viral nucleoproteins (NPs) into helical ribonucleoprotein complexes (RNPs). NP structures of several influenza viruses have been reported. However, there are still contradictory models of how orthomyxovirus RNPs are assembled. Here, we characterize the crystal structure of Thogoto virus (THOV) NP and found striking similarities to structures of influenza viral NPs, including a two-lobed domain architecture, a positively charged RNA-binding cleft, and a tail loop important for trimerization and viral transcription. A low-resolution cryo-electron tomography reconstruction of THOV RNPs elucidates a left-handed double helical assembly. By providing a model for RNP assembly of THOV, our study suggests conserved NP assembly and RNA encapsidation modes for thogoto- and influenza viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19599.map.gz emd_19599.map.gz | 25.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19599-v30.xml emd-19599-v30.xml emd-19599.xml emd-19599.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19599_fsc.xml emd_19599_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_19599.png emd_19599.png | 67.1 KB | ||
| Masks |  emd_19599_msk_1.map emd_19599_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19599.cif.gz emd-19599.cif.gz | 5.6 KB | ||
| Others |  emd_19599_half_map_1.map.gz emd_19599_half_map_1.map.gz emd_19599_half_map_2.map.gz emd_19599_half_map_2.map.gz | 13.3 MB 13.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19599 http://ftp.pdbj.org/pub/emdb/structures/EMD-19599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19599 | HTTPS FTP |
-Validation report
| Summary document |  emd_19599_validation.pdf.gz emd_19599_validation.pdf.gz | 946.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19599_full_validation.pdf.gz emd_19599_full_validation.pdf.gz | 946.4 KB | Display | |
| Data in XML |  emd_19599_validation.xml.gz emd_19599_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_19599_validation.cif.gz emd_19599_validation.cif.gz | 16.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19599 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19599 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19599 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19599 | HTTPS FTP |
-Related structure data
| Related structure data |  8rytMC  8cjwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19599.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19599.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | filtered to 18A, non-sharpened | ||||||||||||||||||||
| Voxel size | X=Y=Z: 2.6 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19599_msk_1.map emd_19599_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19599_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19599_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ribonucleoprotein (RNP) complexes of Thogoto virus
| Entire | Name: Ribonucleoprotein (RNP) complexes of Thogoto virus |
|---|---|
| Components |
|
-Supramolecule #1: Ribonucleoprotein (RNP) complexes of Thogoto virus
| Supramolecule | Name: Ribonucleoprotein (RNP) complexes of Thogoto virus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Thogotovirus Thogotovirus |
-Macromolecule #1: THOV RNP
| Macromolecule | Name: THOV RNP / type: other / ID: 1 / Details: Nucleoprotein / Classification: other |
|---|---|
| Source (natural) | Organism:  Thogotovirus / Strain: THOV strain SiAr 126 Thogotovirus / Strain: THOV strain SiAr 126 |
| Sequence | String: MATDQMDISG PPPKKQHVDT ESQIPKMYEM IRDQMRTLAS THKIPLNIDH NCEVIGSIIM AACTNNRDLR PVDKYWFLMG PAGAEVMTEV EIDIQPQLQW AKGAVHDPKY KGQWYPFLAL LQISNKTKDT ILWQKYPVTQ ELEISNSLEI YANGHGIKDR LKNSRPRSVG ...String: MATDQMDISG PPPKKQHVDT ESQIPKMYEM IRDQMRTLAS THKIPLNIDH NCEVIGSIIM AACTNNRDLR PVDKYWFLMG PAGAEVMTEV EIDIQPQLQW AKGAVHDPKY KGQWYPFLAL LQISNKTKDT ILWQKYPVTQ ELEISNSLEI YANGHGIKDR LKNSRPRSVG PLVHLLHLKR LQENPPKSPA VNGIRKSIVG HLKRQCIGET QKAMINQFEM GRWESLSTFA ASLLAIKPRI ENHFVLTYPL IANCEDFAGA TLSDEWVFKA MEKISNKKTL RVCGPDEKWI SFMNQIYIHS VFQTTGEDLG VLEWVFGGRF CQRKEFGRYC KKSQTKVIGL FTFQYEYWSK PLKSAPRSIE GSKRGQISCR PSFKGKRPSY NNFTSIDALQ SASGSQTVSF YDQVREECQK YMDLKVEGTT CFYRKGGHVE VEFPGSAHCN TYLFG |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 1 / Average exposure time: 1.2 sec. / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 5.0 µm / Calibrated defocus min: 3.0 µm / Calibrated magnification: 33000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 Z
Z Y
Y X
X