[English] 日本語
 Yorodumi
Yorodumi- EMDB-19541: Structure of a yeast 48S-AUC preinitiation complex in closed conf... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a yeast 48S-AUC preinitiation complex in closed conformation | |||||||||
 Map data Map data | For optimal visualization of eIF2 domains (eIFalpha, eIF2gamma and N-terminal domain of eIF2beta), gauss-filter the map by 1.34 and display it at 0.015 contour level, respectively. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / translation / initiation factors / 40S / eIF1A / AUC codon / eIF2 / tRNAi / 48S PIC / small ribosome subunit | |||||||||
| Function / homology |  Function and homology information Function and homology informationformation of translation initiation ternary complex / Recycling of eIF2:GDP / Cellular response to mitochondrial stress / ABC-family proteins mediated transport / methionyl-initiator methionine tRNA binding / translation reinitiation / eukaryotic translation initiation factor 2 complex / formation of cytoplasmic translation initiation complex / multi-eIF complex / protein-synthesizing GTPase ...formation of translation initiation ternary complex / Recycling of eIF2:GDP / Cellular response to mitochondrial stress / ABC-family proteins mediated transport / methionyl-initiator methionine tRNA binding / translation reinitiation / eukaryotic translation initiation factor 2 complex / formation of cytoplasmic translation initiation complex / multi-eIF complex / protein-synthesizing GTPase / eukaryotic 43S preinitiation complex / formation of translation preinitiation complex / eukaryotic 48S preinitiation complex / positive regulation of translational fidelity / Formation of the ternary complex, and subsequently, the 43S complex / Translation initiation complex formation / Ribosomal scanning and start codon recognition / Formation of a pool of free 40S subunits / L13a-mediated translational silencing of Ceruloplasmin expression / ribosomal small subunit binding / 90S preribosome / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / translation regulator activity / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / translation initiation factor binding / translation initiation factor activity / rescue of stalled ribosome / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / translational initiation / protein kinase C binding / positive regulation of apoptotic signaling pathway / modification-dependent protein catabolic process / cytoplasmic stress granule / protein tag activity / rRNA processing / ribosomal small subunit biogenesis / double-stranded RNA binding / small ribosomal subunit rRNA binding / ribosome binding / ribosomal small subunit assembly / small ribosomal subunit / cytosolic small ribosomal subunit / cytoplasmic translation / rRNA binding / ribosome / protein ubiquitination / structural constituent of ribosome / ribonucleoprotein complex / positive regulation of protein phosphorylation / translation / GTPase activity / mRNA binding / ubiquitin protein ligase binding / GTP binding / nucleolus / protein kinase binding / RNA binding / zinc ion binding / nucleus / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) (yeast) / Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) (yeast) /   Kluyveromyces lactis (yeast) / Kluyveromyces lactis (yeast) /  Kluyveromyces lactis NRRL Y-1140 (yeast) / Kluyveromyces lactis NRRL Y-1140 (yeast) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Villamayor-Belinchon L / Sharma P / Llacer JL / Hussain T | |||||||||
| Funding support |  Spain, Spain,  India, 2 items India, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Structural basis of AUC codon discrimination during translation initiation in yeast. Authors: Laura Villamayor-Belinchón / Prafful Sharma / Yuliya Gordiyenko / Jose L Llácer / Tanweer Hussain /    Abstract: In eukaryotic translation initiation, the 48S preinitiation complex (PIC) scans the 5' untranslated region of mRNAs to search for the cognate start codon (AUG) with assistance from various ...In eukaryotic translation initiation, the 48S preinitiation complex (PIC) scans the 5' untranslated region of mRNAs to search for the cognate start codon (AUG) with assistance from various eukaryotic initiation factors (eIFs). Cognate start codon recognition is precise, rejecting near-cognate codons with a single base difference. However, the structural basis of discrimination of near-cognate start codons was not known. We have captured multiple yeast 48S PICs with a near-cognate AUC codon at the P-site, revealing that the AUC codon induces instability in the codon-anticodon at the P-site, leading to a disordered N-terminal tail of eIF1A. Following eIF1 dissociation, the N-terminal domain of eIF5 fails to occupy the vacant eIF1 position, and eIF2β becomes flexible. Consequently, 48S with an AUC codon is less favourable for initiation. Furthermore, we observe hitherto unreported metastable states of the eIF2-GTP-Met-tRNAMet ternary complex, where the eIF2β helix-turn-helix domain may facilitate eIF5 association by preventing eIF1 rebinding to 48S PIC. Finally, a swivelled head conformation of 48S PIC appears crucial for discriminating incorrect and selection of the correct codon-anticodon pair during translation initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19541.map.gz emd_19541.map.gz | 95.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19541-v30.xml emd-19541-v30.xml emd-19541.xml emd-19541.xml | 71.4 KB 71.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19541.png emd_19541.png | 63.8 KB | ||
| Masks |  emd_19541_msk_1.map emd_19541_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19541.cif.gz emd-19541.cif.gz | 14.8 KB | ||
| Others |  emd_19541_half_map_1.map.gz emd_19541_half_map_1.map.gz emd_19541_half_map_2.map.gz emd_19541_half_map_2.map.gz | 91.4 MB 91.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19541 http://ftp.pdbj.org/pub/emdb/structures/EMD-19541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19541 | HTTPS FTP |
-Validation report
| Summary document |  emd_19541_validation.pdf.gz emd_19541_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19541_full_validation.pdf.gz emd_19541_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_19541_validation.xml.gz emd_19541_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_19541_validation.cif.gz emd_19541_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19541 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19541 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19541 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19541 | HTTPS FTP |
-Related structure data
| Related structure data |  8rw1MC  8s8dC  8s8eC  8s8fC  8s8gC  8s8hC  8s8iC  8s8jC  8s8kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19541.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19541.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | For optimal visualization of eIF2 domains (eIFalpha, eIF2gamma and N-terminal domain of eIF2beta), gauss-filter the map by 1.34 and display it at 0.015 contour level, respectively. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||
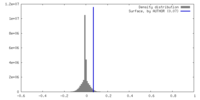
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19541_msk_1.map emd_19541_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
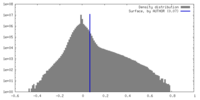
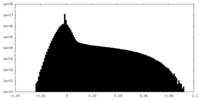
| Density Histograms |
-Half map: #2
| File | emd_19541_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
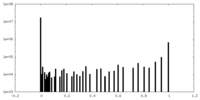
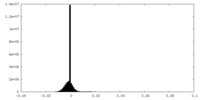
| Density Histograms |
-Half map: #1
| File | emd_19541_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
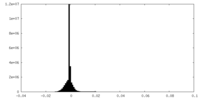
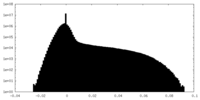
| Density Histograms |
- Sample components
Sample components
+Entire : Structure of a yeast 48S-AUC preinitiation complex in closed conf...
+Supramolecule #1: Structure of a yeast 48S-AUC preinitiation complex in closed conf...
+Supramolecule #2: Ribosome
+Supramolecule #3: tRNA
+Supramolecule #4: Initiation factors eIF1A
+Supramolecule #5: Initiation factor eIF2
+Supramolecule #6: mRNA
+Macromolecule #1: 18S ribosomal RNA
+Macromolecule #37: mRNA (5'-R(P*AP*AP*U)-3')
+Macromolecule #38: Met-tRNAi
+Macromolecule #2: Small ribosomal subunit protein uS2
+Macromolecule #3: Small ribosomal subunit protein eS1
+Macromolecule #4: Small ribosomal subunit protein uS5
+Macromolecule #5: 40S ribosomal protein S4
+Macromolecule #6: Small ribosomal subunit protein eS6
+Macromolecule #7: 40S ribosomal protein S7
+Macromolecule #8: 40S ribosomal protein S8
+Macromolecule #9: KLLA0E23673p
+Macromolecule #10: KLLA0A10483p
+Macromolecule #11: KLLA0F18040p
+Macromolecule #12: Small ribosomal subunit protein uS11
+Macromolecule #13: 40S ribosomal protein S21
+Macromolecule #14: Small ribosomal subunit protein uS8
+Macromolecule #15: KLLA0B11231p
+Macromolecule #16: 40S ribosomal protein S24
+Macromolecule #17: 40S ribosomal protein S26
+Macromolecule #18: 40S ribosomal protein S27
+Macromolecule #19: 40S ribosomal protein S30
+Macromolecule #20: 40S ribosomal protein L41-A
+Macromolecule #21: 40S ribosomal protein S3
+Macromolecule #22: KLLA0D10659p
+Macromolecule #23: KLLA0B08173p
+Macromolecule #24: 40S ribosomal protein S12
+Macromolecule #25: KLLA0F07843p
+Macromolecule #26: Small ribosomal subunit protein uS9
+Macromolecule #27: KLLA0B01474p
+Macromolecule #28: KLLA0B01562p
+Macromolecule #29: KLLA0A07194p
+Macromolecule #30: Small ribosomal subunit protein uS10
+Macromolecule #31: 40S ribosomal protein S25
+Macromolecule #32: Small ribosomal subunit protein eS28
+Macromolecule #33: Small ribosomal subunit protein uS14
+Macromolecule #34: Small ribosomal subunit protein eS31
+Macromolecule #35: KLLA0E12277p
+Macromolecule #36: Eukaryotic translation initiation factor 1A
+Macromolecule #39: Eukaryotic translation initiation factor 2 subunit alpha
+Macromolecule #40: Eukaryotic translation initiation factor 2 subunit gamma
+Macromolecule #41: Eukaryotic translation initiation factor 2 subunit beta
+Macromolecule #42: MAGNESIUM ION
+Macromolecule #43: ZINC ION
+Macromolecule #44: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
+Macromolecule #45: METHIONINE
+Macromolecule #46: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90.0 K / Max: 100.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 8 / Number real images: 11245 / Average exposure time: 1.1 sec. / Average electron dose: 30.0 e/Å2 Details: Images were collected in movie-mode at 30 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
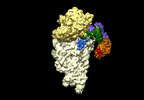
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)