+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | WT-CGS sample in nanodisc | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | cgs / cyclic glucan / ANTIBIOTIC | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglycosyltransferase activity / carbohydrate binding / carbohydrate metabolic process / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) | |||||||||||||||
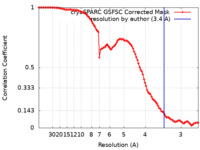
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Sedzicki J / Ni D / Lehmann F / Stahlberg H / Dehio C | |||||||||||||||
| Funding support |  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure-function analysis of the cyclic β-1,2-glucan synthase from Agrobacterium tumefaciens. Authors: Jaroslaw Sedzicki / Dongchun Ni / Frank Lehmann / Henning Stahlberg / Christoph Dehio /  Abstract: The synthesis of complex sugars is a key aspect of microbial biology. Cyclic β-1,2-glucan (CβG) is a circular polysaccharide critical for host interactions of many bacteria, including major ...The synthesis of complex sugars is a key aspect of microbial biology. Cyclic β-1,2-glucan (CβG) is a circular polysaccharide critical for host interactions of many bacteria, including major pathogens of humans (Brucella) and plants (Agrobacterium). CβG is produced by the cyclic glucan synthase (Cgs), a multi-domain membrane protein. So far, its structure as well as the mechanism underlining the synthesis have not been clarified. Here we use cryo-electron microscopy (cryo-EM) and functional approaches to study Cgs from A. tumefaciens. We determine the structure of this complex protein machinery and clarify key aspects of CβG synthesis, revealing a distinct mechanism that uses a tyrosine-linked oligosaccharide intermediate in cycles of polymerization and processing of the glucan chain. Our research opens possibilities for combating pathogens that rely on polysaccharide virulence factors and may lead to synthetic biology approaches for producing complex cyclic sugars. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19114.map.gz emd_19114.map.gz | 97.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19114-v30.xml emd-19114-v30.xml emd-19114.xml emd-19114.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19114_fsc.xml emd_19114_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_19114.png emd_19114.png | 66.3 KB | ||
| Filedesc metadata |  emd-19114.cif.gz emd-19114.cif.gz | 7.3 KB | ||
| Others |  emd_19114_additional_1.map.gz emd_19114_additional_1.map.gz emd_19114_half_map_1.map.gz emd_19114_half_map_1.map.gz emd_19114_half_map_2.map.gz emd_19114_half_map_2.map.gz | 97.3 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19114 http://ftp.pdbj.org/pub/emdb/structures/EMD-19114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19114 | HTTPS FTP |
-Validation report
| Summary document |  emd_19114_validation.pdf.gz emd_19114_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19114_full_validation.pdf.gz emd_19114_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_19114_validation.xml.gz emd_19114_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_19114_validation.cif.gz emd_19114_validation.cif.gz | 23.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19114 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19114 | HTTPS FTP |
-Related structure data
| Related structure data |  8rf0MC  8rf9C  8rfeC  8rfgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19114.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19114.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.316 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_19114_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19114_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19114_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens
| Entire | Name: cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens |
|---|---|
| Components |
|
-Supramolecule #1: cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens
| Supramolecule | Name: cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) |
-Macromolecule #1: Cyclic beta-(1,2)-glucan synthase NdvB
| Macromolecule | Name: Cyclic beta-(1,2)-glucan synthase NdvB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) |
| Molecular weight | Theoretical: 315.448031 KDa |
| Recombinant expression | Organism:  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) |
| Sequence | String: MSFHITPTAA ARDSETKQID HNDSIRASYM TIEELHDAGA ALSRDGADSL PGFMEFDFFE RHRENEKEIL RVYRTTAVDA ENGATITPA AEWLLDNHYV IEEAIQEVRR DFPRKFYRQL PTMTVGGVTI PRVMALGWLY VAHTHSTVSR ENMTALVDGY Q TSQTLQIG ...String: MSFHITPTAA ARDSETKQID HNDSIRASYM TIEELHDAGA ALSRDGADSL PGFMEFDFFE RHRENEKEIL RVYRTTAVDA ENGATITPA AEWLLDNHYV IEEAIQEVRR DFPRKFYRQL PTMTVGGVTI PRVMALGWLY VAHTHSTVSR ENMTALVDGY Q TSQTLQIG ELWALPSIIR FVLIENLRRI SIRVERSRRM RQKANEVVDE IIRLNDAEAS AALLKQVDSL VDDPTFATQF LY RLRNGSQ TSGFAVAWLE ERLHAAGTDA ENVMMSEHNR LASGNVTMGN IVKSLREIDD TEWSVWFEEV SHIDKVLREE TDY EILDFG SRNTYRNTIE LLARRSPRTE VEVARAAVEM ARSDLPAGAD ENHRVNVGSV LVGQRRFELE KALGYRPLAS QHIV RSMRK FNWLAIAAPV LLITAVAMLA VGWFLAKAGM PWYVVTAFLL MFALPASEGA TGLFNTLVTF FVKPFRLVGY EFKNG IPED ARTLVAVPCM LTSRDSVDEM MRNIEVHYLA NPHGEIYFSL VSDWRDAPYE QSDEDLEILD YAKRELAALN SRYAFD GKT RFYLLHRRRI YNSAEECWMG WERKRGKLHE LNMLLRGDKD TTFLGGSNIV PADVKYVMTL DADTRLMRDA VTKLVGK MH HPINRPKIDP VSGRVVEGYG LLQPRVTPSL TTGKDASVFQ RVFSINRGID PYVFTVSDVY QDLTSEGTFT GKGLYDVD A FEAALKGRIE ENSILSHDLL EGSFARCALV TDVELVEDFP TRYEVEVSRQ HRWARGDWQL LPFIIDRARG VTAIGRWKM VDNLRRSLTP IAWFFASILG WYFMDPLGAL IWQILLIFSL FVAPTLSLLS GLVPRSTDIV PQAHFFTIWS EIRATNAQVA LRIVFIADA ACMMTDAIVR SLYRLLVSHK LMLEWRTAAS MQSSAQGSIV DYYRQMWHAP VVAMLGLLFA ALPGDNAFLI G IPFTLLWV LSPAVAWYVS QSAETEDRLF VSEHVSFELR KIARRTWRYY EAFVTPQENH LPPDNFQETP EPIVASRTSP TN IGVYLLS VISARQFGWI SFADTLERIE NTIQTVEKME KHRGHLYNWY HTDTLQTLGP RYVSAVDSGN LAGHLIAVSS ACR DWAEAP SAHLQGNLDG IGDVAGILRE TLKALPDNRK TLRPLHRRLE ERIIGFSNAL ASVKREHEFA SIRVINLAVL ARDI QKLAT NVDHEVKSAQ SAEVTRWAQL LVESCEAHIS DSAIDLTNME PLRQRLASLR DRSRNLAFSM DFTFLYRKDR RLLSI GYRV ESKELDEACY DLLASECRLT SLFAIAKGDL PTEHWYRLGR QVVPIGAQGA LVSWSGSMFE YLMPPLVMQE RQGGIL NQT NNLIVKEQMN HGRRLGTPWG ISEAAFNARD HNMNYQYTNF GVPTLGLKRG LGQNAVIAPY ASILASQYDP DGALENL DK LRKLGALGQY GFHDAVDFTP TRVPDGKVCA VVYNYYAHHH GMSIAAVANV AFDGVLRELF HSDPVIEAAE LLLQEKAP R EVPVMSAKYE PETPGKEQAD LLRAEVRSIA DPAVRDREVV FLSNGHYSTM LTSTGAGYSK WNGQAISRWK ADPTDDRWG TFIFLRDTTN GQWWSATAEP RVIEGEKTKT IFTDDKAEFH KTIGDLQSVV ECIVATEHDA EGRRITLLNV GSEDRYIEVT SYMEPVIAS EDDDNAHPLF SRMFVQTEIG RRGDVIRAWR NRRSQNEPGT VIAHLAADNA GPSRPTEFET DRAKFIGRGR S LREAAAFD AGATLSSSDG FTLDPILSLR RTVRVPAGKK VSVIFWTIAA PSREEVDKAI DRYRHPDAFA HELVHAWTRT QV QMRHVGV TSQQAAAFQH LGRYLTYPDM HLRADSETLK TGLASQRALW PLAISGDFPI FSLRINDDMD MDIAREALSA HEY LRSRGV IFDLVIVNER AASYAQDMQH ALDHISETQR RINPADGGRP HVFSVRRDLM DEETWSALLA ASRVVLHVRN GKIV DQINR AVSLFAANRG PDGSSDAAQA RLPVPAFPVA EPVEDAGDLD FWNGFGGFAK NGQEYVVRLN GGQSTPHPWI NVISN ENFG FHISAEGAGF SWSRNSRDYQ LTPWTNDPVI NRPGEAFYVA DVETGKLYTP CAALSRDPEA MFETRHGLGY SILTGV ADT LEVELTQTVD REKPVKFSQV IVRNKGSKSR RLKVYAYVEW VLGNNGQKSA PFILSRHDAG SNAIFASNPY SIDYSAR TS FLTLDSEASG FTTSRREFIG RFGSAQAPQG IVAGAALSGT TEVDGDPCAA LMQEIHLKPG EERHMTFILG DADNAEEA E ALVKDVRQAD FLSVLEESKK FWTGFTGQLQ VSTPDAGFNH MVNNWLPYQA LACRILARTA FYQSSGAFGF RDQLQDTLA FLLYQPDLAR TQILRAAGRQ FPEGDVQHWW LPLTGAGVRT TISDDVVWLA YAINQYVSAT GDAAILDESI PFLKGPALMP GQHDAFFQP ETSERSATLY EHAALALDLA IHRTGENGLP LILGGDWNDG MNRVGVGGKG TSVWLGWFLA GALRDFIEIA E KRGDTDRV GKWASHREKL RHVLETAGWD GSYYRRGYFD DGTPLGSASS EECQIDSLGQ SWSVLSGEGE EGRSRQAMDA VM EHLVDEK TGIIRLFTPP FSRASHDPGY IKGYPPGVRE NGGQYTHAAT WVVLALAKQG RAQEAWNCFK LLNPVNHALD AAS SETYRV EPYVVTADVY GEGAYAGRGG WSWYTGSAGW LYRAAVEGIL GITRTDGKLH VSPSLPEDWS GFSIRITLDG KARD IAVSR KAGTADVSVS VDG UniProtKB: Cyclic beta-(1,2)-glucan synthase NdvB |
-Macromolecule #4: URIDINE-5'-DIPHOSPHATE-GLUCOSE
| Macromolecule | Name: URIDINE-5'-DIPHOSPHATE-GLUCOSE / type: ligand / ID: 4 / Number of copies: 1 / Formula: UPG |
|---|---|
| Molecular weight | Theoretical: 566.302 Da |
| Chemical component information | 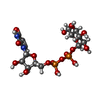 ChemComp-UPG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)