[English] 日本語
 Yorodumi
Yorodumi- EMDB-19055: Escherichia coli paused disome complex (Non-rotated disome interf... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Escherichia coli paused disome complex (Non-rotated disome interface class 2) | |||||||||
 Map data Map data | Cryo-EM reconstruction of the E. coli disome complex (non-rotated disome interface class 2). Full map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / polysome / tranlsation / elongation / pausing / disome / collision / PURE system | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA secondary structure unwinding / positive regulation of cytoplasmic translation / negative regulation of cytoplasmic translational initiation / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / positive regulation of ribosome biogenesis / negative regulation of cytoplasmic translation ...RNA secondary structure unwinding / positive regulation of cytoplasmic translation / negative regulation of cytoplasmic translational initiation / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / positive regulation of ribosome biogenesis / negative regulation of cytoplasmic translation / four-way junction DNA binding / DnaA-L2 complex / negative regulation of DNA-templated DNA replication initiation / negative regulation of translational initiation / regulation of mRNA stability / mRNA regulatory element binding translation repressor activity / assembly of large subunit precursor of preribosome / positive regulation of RNA splicing / transcription elongation factor complex / cytosolic ribosome assembly / regulation of DNA-templated transcription elongation / DNA endonuclease activity / transcription antitermination / regulation of cell growth / translational initiation / DNA-templated transcription termination / maintenance of translational fidelity / mRNA 5'-UTR binding / ribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / ribosome biogenesis / ribosome binding / regulation of translation / ribosomal small subunit assembly / small ribosomal subunit / 5S rRNA binding / large ribosomal subunit rRNA binding / transferase activity / cytosolic small ribosomal subunit / ribosomal large subunit assembly / cytoplasmic translation / cytosolic large ribosomal subunit / tRNA binding / molecular adaptor activity / negative regulation of translation / single-stranded RNA binding / rRNA binding / ribosome / structural constituent of ribosome / translation / response to antibiotic / mRNA binding / RNA binding / zinc ion binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
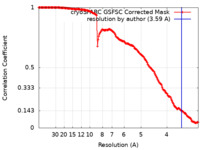
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Fluegel T / Schacherl M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Transient disome complex formation in native polysomes during ongoing protein synthesis captured by cryo-EM. Authors: Timo Flügel / Magdalena Schacherl / Anett Unbehaun / Birgit Schroeer / Marylena Dabrowski / Jörg Bürger / Thorsten Mielke / Thiemo Sprink / Christoph A Diebolder / Yollete V Guillén ...Authors: Timo Flügel / Magdalena Schacherl / Anett Unbehaun / Birgit Schroeer / Marylena Dabrowski / Jörg Bürger / Thorsten Mielke / Thiemo Sprink / Christoph A Diebolder / Yollete V Guillén Schlippe / Christian M T Spahn /  Abstract: Structural studies of translating ribosomes traditionally rely on in vitro assembly and stalling of ribosomes in defined states. To comprehensively visualize bacterial translation, we reactivated ex ...Structural studies of translating ribosomes traditionally rely on in vitro assembly and stalling of ribosomes in defined states. To comprehensively visualize bacterial translation, we reactivated ex vivo-derived E. coli polysomes in the PURE in vitro translation system and analyzed the actively elongating polysomes by cryo-EM. We find that 31% of 70S ribosomes assemble into disome complexes that represent eight distinct functional states including decoding and termination intermediates, and a pre-nucleophilic attack state. The functional diversity of disome complexes together with RNase digest experiments suggests that paused disome complexes transiently form during ongoing elongation. Structural analysis revealed five disome interfaces between leading and queueing ribosomes that undergo rearrangements as the leading ribosome traverses through the elongation cycle. Our findings reveal at the molecular level how bL9's CTD obstructs the factor binding site of queueing ribosomes to thwart harmful collisions and illustrate how translation dynamics reshape inter-ribosomal contacts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19055.map.gz emd_19055.map.gz | 348.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19055-v30.xml emd-19055-v30.xml emd-19055.xml emd-19055.xml | 67.3 KB 67.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19055_fsc.xml emd_19055_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_19055.png emd_19055.png | 67.5 KB | ||
| Masks |  emd_19055_msk_1.map emd_19055_msk_1.map | 391 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19055.cif.gz emd-19055.cif.gz | 14.9 KB | ||
| Others |  emd_19055_half_map_1.map.gz emd_19055_half_map_1.map.gz emd_19055_half_map_2.map.gz emd_19055_half_map_2.map.gz | 362.4 MB 362.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19055 http://ftp.pdbj.org/pub/emdb/structures/EMD-19055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19055 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19055 | HTTPS FTP |
-Validation report
| Summary document |  emd_19055_validation.pdf.gz emd_19055_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19055_full_validation.pdf.gz emd_19055_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_19055_validation.xml.gz emd_19055_validation.xml.gz | 24.9 KB | Display | |
| Data in CIF |  emd_19055_validation.cif.gz emd_19055_validation.cif.gz | 32.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19055 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19055 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19055 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19055 | HTTPS FTP |
-Related structure data
| Related structure data |  8rcmMC  8pegC  8pklC  8r3vC  8rclC  8rcsC  8rctC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19055.map.gz / Format: CCP4 / Size: 391 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19055.map.gz / Format: CCP4 / Size: 391 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the E. coli disome complex (non-rotated disome interface class 2). Full map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.59 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19055_msk_1.map emd_19055_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM reconstruction of the E. coli disome complex...
| File | emd_19055_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the E. coli disome complex (non-rotated disome interface class 2). Half map A. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM reconstruction of the E. coli disome complex...
| File | emd_19055_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the E. coli disome complex (non-rotated disome interface class 2). Half map B. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Escherichia coli paused disome complex
+Supramolecule #1: Escherichia coli paused disome complex
+Macromolecule #1: 50S ribosomal protein L28
+Macromolecule #2: 50S ribosomal protein L30
+Macromolecule #3: Large ribosomal subunit protein bL31A
+Macromolecule #4: 50S ribosomal protein L35
+Macromolecule #8: Small ribosomal subunit protein uS2
+Macromolecule #9: Small ribosomal subunit protein uS3
+Macromolecule #10: Small ribosomal subunit protein uS4
+Macromolecule #11: Small ribosomal subunit protein uS5
+Macromolecule #12: 30S ribosomal protein S6
+Macromolecule #13: 30S ribosomal protein S7
+Macromolecule #14: Small ribosomal subunit protein uS8
+Macromolecule #15: Small ribosomal subunit protein uS9
+Macromolecule #16: 30S ribosomal protein S10
+Macromolecule #17: Small ribosomal subunit protein uS11
+Macromolecule #18: Small ribosomal subunit protein uS12
+Macromolecule #19: Small ribosomal subunit protein uS13
+Macromolecule #20: Small ribosomal subunit protein uS14
+Macromolecule #21: 30S ribosomal protein S15
+Macromolecule #22: 30S ribosomal protein S16
+Macromolecule #23: Small ribosomal subunit protein uS17
+Macromolecule #24: Small ribosomal subunit protein bS18
+Macromolecule #25: Small ribosomal subunit protein uS19
+Macromolecule #26: Small ribosomal subunit protein bS20
+Macromolecule #27: 30S ribosomal protein S21
+Macromolecule #34: 30S ribosomal protein S1
+Macromolecule #35: Large ribosomal subunit protein uL1
+Macromolecule #36: 50S ribosomal protein L2
+Macromolecule #37: 50S ribosomal protein L5
+Macromolecule #38: 50S ribosomal protein L33
+Macromolecule #39: 50S ribosomal protein L16
+Macromolecule #40: 50S ribosomal protein L9
+Macromolecule #41: 50S ribosomal protein L34
+Macromolecule #42: 50S ribosomal protein L15
+Macromolecule #43: Nascent chain
+Macromolecule #44: 50S ribosomal protein L18
+Macromolecule #45: 50S ribosomal protein L27
+Macromolecule #5: 23S ribosomal RNA
+Macromolecule #6: 5S ribosomal RNA
+Macromolecule #7: 16S ribosomal RNA
+Macromolecule #28: messenger RNA
+Macromolecule #29: tRNA-Trp (P-site)
+Macromolecule #30: tRNA-Phe (P-site)
+Macromolecule #31: tRNA-Arg (E-site)
+Macromolecule #32: tRNA-Val (A-site)
+Macromolecule #33: tRNA-Ala (A-site)
+Macromolecule #46: ZINC ION
+Macromolecule #47: 1,4-DIAMINOBUTANE
+Macromolecule #48: MAGNESIUM ION
+Macromolecule #49: ALANINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.037 kPa Details: Operated at 15 mA, easiGlow Discharge Cleaning system (Pelco) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 75 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Withdrawn samples were spotted directly onto freshly glow-discharged holey carbon grids, blotted for 1-2 s, and flash frozen in liquid ethane using a Vitrobot Mark IV plunger (ThermoFisher ...Details: Withdrawn samples were spotted directly onto freshly glow-discharged holey carbon grids, blotted for 1-2 s, and flash frozen in liquid ethane using a Vitrobot Mark IV plunger (ThermoFisher Scientific) after a wait time of 40 s at 4 degrees Celcius.. |
| Details | In vitro translation reactions were performed in the PURE translation system using the PURExpress delta ribosome kit (NEB, #E3313S). Translation reactions were supplemented with 0.8 U/uL RNAsin Plus RNase Inhibitor (Promega, N261B). SolA, factor mix, and RNAsin Plus were combined on ice, followed by a preincubation at 37 degrees Celcius for 2 min, and added directly to polysomes (700 nM final concentration) that had been preincubated at 37 degrees Celsius for 2 min. After 1 min reaction time, 4 uL of the reaction mixture were withdrawn for plunge freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 82.0 K |
| Software | Name: EPU (ver. 2.8.1) |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 8982 / Average exposure time: 1.13 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Software | Name:  Coot (ver. 0.9.6) Coot (ver. 0.9.6) |
| Details | The atomic model of the E. coli 70S ribosome (PDB ID: 7N1P) was initially docked into the postprocessed density maps with UCSF Chimera and manually adjusted in Coot. All models were refined over multiple rounds using the module phenix.real_space_refine in PHENIX and interactive model building and refinement in Coot, using libG restraints for the RNAs. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-8rcm: |
 Movie
Movie Controller
Controller


















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)