+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Capsid structure of Giardiavirus (GLV) HP strain | ||||||||||||
 Map data Map data | Full 3D reconstruction of GLV-HP capsid. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Giardia parasite / dsRNA virus / capsid structure / cryo / Totiviridae / Giardia lambia virus (GLV) / VIRUS | ||||||||||||
| Function / homology | Capsid protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Giardia lamblia virus Giardia lamblia virus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.14 Å | ||||||||||||
 Authors Authors | Wang H / Gianluca M / Munke A / Hassan MM / Lalle M / Okamoto K | ||||||||||||
| Funding support |  Sweden, Sweden,  Norway, 3 items Norway, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Capsid structure of Giardiavirus (GLV) HP strain Authors: Wang H / Gianluca M / Munke A / Hassan MM / Lalle M / Okamoto K | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18791.map.gz emd_18791.map.gz | 936.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18791-v30.xml emd-18791-v30.xml emd-18791.xml emd-18791.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18791.png emd_18791.png | 249.5 KB | ||
| Filedesc metadata |  emd-18791.cif.gz emd-18791.cif.gz | 5.9 KB | ||
| Others |  emd_18791_half_map_1.map.gz emd_18791_half_map_1.map.gz emd_18791_half_map_2.map.gz emd_18791_half_map_2.map.gz | 925 MB 925 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18791 http://ftp.pdbj.org/pub/emdb/structures/EMD-18791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18791 | HTTPS FTP |
-Validation report
| Summary document |  emd_18791_validation.pdf.gz emd_18791_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18791_full_validation.pdf.gz emd_18791_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_18791_validation.xml.gz emd_18791_validation.xml.gz | 22.1 KB | Display | |
| Data in CIF |  emd_18791_validation.cif.gz emd_18791_validation.cif.gz | 26.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18791 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18791 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18791 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18791 | HTTPS FTP |
-Related structure data
| Related structure data |  8r0fMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18791.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18791.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full 3D reconstruction of GLV-HP capsid. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A of the 3D reconstruction of GLV-HP capsid.
| File | emd_18791_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of the 3D reconstruction of GLV-HP capsid. | ||||||||||||
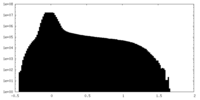
| Projections & Slices |
| ||||||||||||
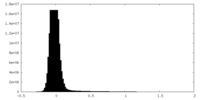
| Density Histograms |
-Half map: Half map B of the 3D reconstruction of GLV-HP capsid.
| File | emd_18791_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of the 3D reconstruction of GLV-HP capsid. | ||||||||||||
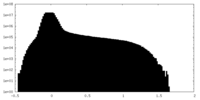
| Projections & Slices |
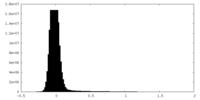
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Giardia lamblia virus
| Entire | Name:  Giardia lamblia virus Giardia lamblia virus |
|---|---|
| Components |
|
-Supramolecule #1: Giardia lamblia virus
| Supramolecule | Name: Giardia lamblia virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: GLV isolated from Giardia duodenalis isolates HP-1 from a human patient. NCBI-ID: 29255 / Sci species name: Giardia lamblia virus / Sci species strain: HP-1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 450.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Giardia lamblia virus Giardia lamblia virus |
| Molecular weight | Theoretical: 103.48132 KDa |
| Sequence | String: PENITFDTLN TQNDHEETYG ESPEVPKASI APAGRQNVPV LQQNQENEDN HALGGSEDAK DEREIQSSAI KTLYNYYSEG PSTPIMPHL VNRLRGLDAL AKVDATLSKV DMNAAYIFAL RPTFPYSYGY KQRFSNRRLT TSALCYARTG LSSFLTVDKT Y TSNSPLKG ...String: PENITFDTLN TQNDHEETYG ESPEVPKASI APAGRQNVPV LQQNQENEDN HALGGSEDAK DEREIQSSAI KTLYNYYSEG PSTPIMPHL VNRLRGLDAL AKVDATLSKV DMNAAYIFAL RPTFPYSYGY KQRFSNRRLT TSALCYARTG LSSFLTVDKT Y TSNSPLKG GSRGWPIFNV GVSPHVAEPH MRTLSPIGLE VFNLATSQFS KTLLTASSKV FTQSLYTADI LSIFGEVFLP HV MQPVSNY TPILVRALLA LIHILGSGSG NCSLSSSIFE SSIPQFLTIS HSTNMSNRTR YCLHTWSAYK DMFRNGIPPQ STF PPTLAP EGSSARILIP AALVTSPMFP WLLVLVSSGP QFFLYSKDAS INTVDIGSRG RITSPIPDVA HLDLHRLWNL FRFD GYRYI DVVIVGVDRD YVWPYQNGVY VHGGKGPKGT DNYENADVHD GIGTIFSSFN NNVNVQTSDL LLGLSTLWNH ITTTY ATEE EVTMAIKIAA AFALVYPVQP IVYSGCSRAL YNHTSYFQPS SENCYTTDTA EVKSTWDTVE LSVQVNNAMV LGMTLP FGQ PTVSSAQWFN NIDKAEISMF KVGNLPLQNL DYLSLDMMEF YAPTTGQLYD IRSDSLISSA HRTVNLGIGY TALADFF AY LASVPAQSFY HNRMVTSPIS KQAYSVYERF IERFIDDFVG WGRCDLFNLD TLLGAKRIAG VASSPIPWHC SLQRCPLP I IMHYTGLHFG QEHIRVRDVA GVEGLQQIVL RNDQGSIVLD ALGTAAPSRL AVKLDWSRLS AWYSDTTCAI PISDRVMEI VNYAAIWDPT QERRATGFVY TYFSPNFLSS FNVSEPIFNK TINLTPPYDD TSQTVIQNLS MPQMLSFDPY YESTFYVVSA DNEWVPTSG PAWKVPYLEN VVKRSGRRLL AELRIASNNG SGDRTFLDDV UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 1 x PBS (-) |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified GLV-HP particles |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.14 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 28342 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X