[English] 日本語
 Yorodumi
Yorodumi- EMDB-18784: CryoEM structure of DHS-eIF5A complex structure from Trichomonas ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
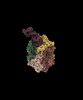
| Title | CryoEM structure of DHS-eIF5A complex structure from Trichomonas vaginalis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | parasite / hypusination / eI5FA / hypusine / DHS / Trichomonas vaginalis / deoxyhypusination / translation factor / TRANSLATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationdeoxyhypusine synthase activity / positive regulation of translational termination / positive regulation of translational elongation / translation elongation factor activity / translation initiation factor activity / ribosome binding / RNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Trichomonas vaginalis (eukaryote) Trichomonas vaginalis (eukaryote) | |||||||||
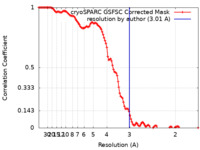
| Method | single particle reconstruction / cryo EM / Resolution: 3.01 Å | |||||||||
 Authors Authors | Wator E / Wilk P / Grudnik P | |||||||||
| Funding support |  Poland, 2 items Poland, 2 items
| |||||||||
 Citation Citation |  Journal: FEBS J / Year: 2024 Journal: FEBS J / Year: 2024Title: Structural characterization of the (deoxy)hypusination in Trichomonas vaginalis questions the bifunctionality of deoxyhypusine synthase. Authors: Elżbieta Wątor / Piotr Wilk / Paweł Kochanowski / Przemysław Grudnik /  Abstract: Trichomonas vaginalis, the causative agent of trichomoniasis, is a prevalent anaerobic protozoan parasite responsible for the most common nonviral sexually transmitted infection globally. While ...Trichomonas vaginalis, the causative agent of trichomoniasis, is a prevalent anaerobic protozoan parasite responsible for the most common nonviral sexually transmitted infection globally. While metronidazole and its derivatives are approved drugs for this infection, rising resistance necessitates the exploration of new antiparasitic therapies. Protein posttranslational modifications (PTMs) play crucial roles in cellular processes, and among them, hypusination, involving eukaryotic translation factor 5A (eIF5A), has profound implications. Despite extensive studies in various organisms, the role of hypusination in T. vaginalis and its potential impact on parasite biology and pathogenicity remain poorly understood. This study aims to unravel the structural basis of the hypusination pathway in T. vaginalis using X-ray crystallography and cryo-electron microscopy. The results reveal high structural homology between T. vaginalis and human orthologs, providing insights into the molecular architecture of eIF5A and deoxyhypusine synthase (DHS) and their interaction. Contrary to previous suggestions of bifunctionality, our analyses indicate that the putative hydroxylation site in tvDHS is nonfunctional, and biochemical assays demonstrate exclusive deoxyhypusination capability. These findings challenge the notion of tvDHS functioning as both deoxyhypusine synthase and hydroxylase. The study enhances understanding of the hypusination pathway in T. vaginalis, shedding light on its functional relevance and potential as a drug target, and contributing to the development of novel therapeutic strategies against trichomoniasis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18784.map.gz emd_18784.map.gz | 157 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18784-v30.xml emd-18784-v30.xml emd-18784.xml emd-18784.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18784_fsc.xml emd_18784_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18784.png emd_18784.png | 39.5 KB | ||
| Filedesc metadata |  emd-18784.cif.gz emd-18784.cif.gz | 6.3 KB | ||
| Others |  emd_18784_half_map_1.map.gz emd_18784_half_map_1.map.gz emd_18784_half_map_2.map.gz emd_18784_half_map_2.map.gz | 154.4 MB 154.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18784 http://ftp.pdbj.org/pub/emdb/structures/EMD-18784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18784 | HTTPS FTP |
-Related structure data
| Related structure data |  8qzxMC  8qzvC  8qzwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18784.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18784.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8456 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18784_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18784_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of DHS-eIF5A
| Entire | Name: Complex of DHS-eIF5A |
|---|---|
| Components |
|
-Supramolecule #1: Complex of DHS-eIF5A
| Supramolecule | Name: Complex of DHS-eIF5A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Trichomonas vaginalis (eukaryote) Trichomonas vaginalis (eukaryote) |
-Macromolecule #1: Deoxyhypusine synthase related protein, putative
| Macromolecule | Name: Deoxyhypusine synthase related protein, putative / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Trichomonas vaginalis (eukaryote) Trichomonas vaginalis (eukaryote) |
| Molecular weight | Theoretical: 40.489102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRAPSLAPS LAVDSVFVES EELTTPLVKG YVPDDNGKFD FDKMLEQMKY CGFQATNLGL AIDQINEMLH YDYEPQPGDE KKLFGLGGG VEGVKYKPRA CKIFLGITSN LISSGMRDYI RFLVKHALVD VVVCTAGGIE EDFIKCLAPT HMGEFFHDGH D LRKRGLNR ...String: MSRAPSLAPS LAVDSVFVES EELTTPLVKG YVPDDNGKFD FDKMLEQMKY CGFQATNLGL AIDQINEMLH YDYEPQPGDE KKLFGLGGG VEGVKYKPRA CKIFLGITSN LISSGMRDYI RFLVKHALVD VVVCTAGGIE EDFIKCLAPT HMGEFFHDGH D LRKRGLNR IGNLIVPNKN YCLFEDWIMP ILDKCLEEQN TQGTKWTPSK LIHRLGLEIN NEDSVWYWAA KNNIPVYSPA LT DGSIGDM IYFHSYNNPG LVLDLVEDIR DMNNEPLWAT KTGCIILGGG VVKHHIMNAN LYRNGADFVV YVNTAHDFDG SDS GARPDE AVSWGAISLE AKPVKVYAEV TLVLPLLVAG SFSKFLAE UniProtKB: Deoxyhypusine synthase related protein, putative |
-Macromolecule #2: Eukaryotic translation initiation factor 5A
| Macromolecule | Name: Eukaryotic translation initiation factor 5A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Trichomonas vaginalis (eukaryote) Trichomonas vaginalis (eukaryote) |
| Molecular weight | Theoretical: 18.430594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSAEEEVHH DLEIQEVDAG SQEKATIPVN KLKKGGYVLI EGRPCRVVDI TKSKTGKHGH AKAGIAGTDL FTGRRYETHL PTSHEIEVP FVDRSDYGLI NIDDGHTQLL TLDGTLREDV DLPPEGNEMR QRVIDLFNVC VNTNDQVVVT VLSSNGENLI V DCKKSTN UniProtKB: Eukaryotic translation initiation factor 5A |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 4 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Macromolecule #4: SPERMIDINE
| Macromolecule | Name: SPERMIDINE / type: ligand / ID: 4 / Number of copies: 3 / Formula: SPD |
|---|---|
| Molecular weight | Theoretical: 145.246 Da |
| Chemical component information |  ChemComp-SPD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 9.3 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 70 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)