[English] 日本語
 Yorodumi
Yorodumi- EMDB-18455: Human Adenovirus type 11 fiber knob in complex with its cell rece... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Adenovirus type 11 fiber knob in complex with its cell receptors, Desmoglein-2 and CD46 | |||||||||
 Map data Map data | This is the 3D reconstruction of hAd11k in complex with rDSG2 and CD46 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adenovirus / cell entry / receptor binding / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsequestering of extracellular ligand from receptor / inner acrosomal membrane / Purkinje myocyte development / bundle of His cell-Purkinje myocyte adhesion involved in cell communication / cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication / desmosome organization / Keratinization / regulation of Notch signaling pathway / desmosome / negative regulation of complement activation, classical pathway ...sequestering of extracellular ligand from receptor / inner acrosomal membrane / Purkinje myocyte development / bundle of His cell-Purkinje myocyte adhesion involved in cell communication / cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication / desmosome organization / Keratinization / regulation of Notch signaling pathway / desmosome / negative regulation of complement activation, classical pathway / positive regulation of transforming growth factor beta production / T cell mediated immunity / Formation of the cornified envelope / cornified envelope / positive regulation of memory T cell differentiation / regulation of ventricular cardiac muscle cell action potential / positive regulation of regulatory T cell differentiation / Apoptotic cleavage of cell adhesion proteins / adhesion receptor-mediated virion attachment to host cell / regulation of heart rate by cardiac conduction / homophilic cell adhesion via plasma membrane adhesion molecules / positive regulation of interleukin-10 production / intercalated disc / RHOG GTPase cycle / lateral plasma membrane / single fertilization / RAC2 GTPase cycle / RAC3 GTPase cycle / maternal process involved in female pregnancy / positive regulation of T cell proliferation / cell adhesion molecule binding / complement activation, classical pathway / Regulation of Complement cascade / response to progesterone / cell-cell adhesion / viral capsid / cell-cell junction / cell junction / virus receptor activity / signaling receptor activity / adaptive immune response / cell adhesion / cadherin binding / symbiont entry into host cell / apical plasma membrane / negative regulation of gene expression / intracellular membrane-bounded organelle / innate immune response / focal adhesion / calcium ion binding / host cell nucleus / positive regulation of gene expression / cell surface / extracellular space / extracellular exosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Human adenovirus 11 / Human adenovirus 11 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
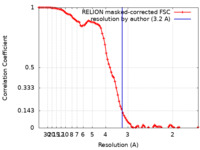
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Effantin G | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2023 Journal: J Virol / Year: 2023Title: Toward the understanding of DSG2 and CD46 interaction with HAdV-11 fiber, a super-complex analysis. Authors: Gregory Effantin / Marc-André Hograindleur / Daphna Fenel / Pascal Fender / Emilie Vassal-Stermann /  Abstract: The main limitation of oncolytic vectors is neutralization by blood components, which prevents intratumoral administration to patients. Enadenotucirev, a chimeric HAdV-11p/HAdV-3 adenovirus ...The main limitation of oncolytic vectors is neutralization by blood components, which prevents intratumoral administration to patients. Enadenotucirev, a chimeric HAdV-11p/HAdV-3 adenovirus identified by bio-selection, is a low seroprevalence vector active against a broad range of human carcinoma cell lines. At this stage, there's still some uncertainty about tropism and primary receptor utilization by HAdV-11. However, this information is very important, as it has a direct influence on the effectiveness of HAdV-11-based vectors. The aim of this work is to determine which of the two receptors, DSG2 and CD46, is involved in the attachment of the virus to the host, and what role they play in the early stages of infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18455.map.gz emd_18455.map.gz | 46.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18455-v30.xml emd-18455-v30.xml emd-18455.xml emd-18455.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18455_fsc.xml emd_18455_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18455.png emd_18455.png | 92.1 KB | ||
| Masks |  emd_18455_msk_1.map emd_18455_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18455.cif.gz emd-18455.cif.gz | 6.6 KB | ||
| Others |  emd_18455_half_map_1.map.gz emd_18455_half_map_1.map.gz emd_18455_half_map_2.map.gz emd_18455_half_map_2.map.gz | 46.3 MB 46.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18455 http://ftp.pdbj.org/pub/emdb/structures/EMD-18455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18455 | HTTPS FTP |
-Validation report
| Summary document |  emd_18455_validation.pdf.gz emd_18455_validation.pdf.gz | 827.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18455_full_validation.pdf.gz emd_18455_full_validation.pdf.gz | 827.1 KB | Display | |
| Data in XML |  emd_18455_validation.xml.gz emd_18455_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_18455_validation.cif.gz emd_18455_validation.cif.gz | 19.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18455 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18455 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18455 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18455 | HTTPS FTP |
-Related structure data
| Related structure data |  8qk3MC  8qjxC  8qjyC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18455.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18455.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D reconstruction of hAd11k in complex with rDSG2 and CD46 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18455_msk_1.map emd_18455_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: This is the 3D reconstruction of hAd11k in...
| File | emd_18455_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D reconstruction of hAd11k in complex with rDSG2 and CD46 - half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: This is the 3D reconstruction of hAd11k in...
| File | emd_18455_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D reconstruction of hAd11k in complex with rDSG2 and CD46 - half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between the human adenovirus 11 fiber knob, the human des...
| Entire | Name: Complex between the human adenovirus 11 fiber knob, the human desmoglein 2 (domains ec2/ec3) and the human CD46 (domains SCR1 and SCR2) |
|---|---|
| Components |
|
-Supramolecule #1: Complex between the human adenovirus 11 fiber knob, the human des...
| Supramolecule | Name: Complex between the human adenovirus 11 fiber knob, the human desmoglein 2 (domains ec2/ec3) and the human CD46 (domains SCR1 and SCR2) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Fiber protein
| Macromolecule | Name: Fiber protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human adenovirus 11 Human adenovirus 11 |
| Molecular weight | Theoretical: 35.564773 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKRVRLSDS FNPVYPYEDE STSQHPFINP GFISPNGFTQ SPNGVLTLKC LTPLTTTGGS LQLKVGGGLT VDDTNGFLKE NISATTPLV KTGHSIGLPL GAGLGTNENK LCIKLGQGLT FNSNNICIDD NINTLWTGVN PTEANCQIMN SSESNDCKLI L TLVKTGAL ...String: MTKRVRLSDS FNPVYPYEDE STSQHPFINP GFISPNGFTQ SPNGVLTLKC LTPLTTTGGS LQLKVGGGLT VDDTNGFLKE NISATTPLV KTGHSIGLPL GAGLGTNENK LCIKLGQGLT FNSNNICIDD NINTLWTGVN PTEANCQIMN SSESNDCKLI L TLVKTGAL VTAFVYVIGV SNNFNMLTTH RNINFTAELF FDSTGNLLTR LSSLKTPLNH KSGQNMATGA ITNAKGFMPS TT AYPFNDN SREKENYIYG TCYYTASDRT AFPIDISVML NRRAINDETS YCIRITWSWN TGDAPEVQTS ATTLVTSPFT FYY IREDD UniProtKB: Fiber protein |
-Macromolecule #2: Membrane cofactor protein
| Macromolecule | Name: Membrane cofactor protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.795164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEPPGRRECP FPSWRFPGLL LAAMVLLLYS FSDACEEPPT FEAMELIGKP KPYYEIGERV DYKCKKGYFY IPPLATHTIC DRNHTWLPV SDDACYRETC PYIRDPLNGQ AVPANGTYEF GYQMHFICNE GYYLIGEEIL YCELKGSVAI WSGKPPICEK V LCTPPPKI ...String: MEPPGRRECP FPSWRFPGLL LAAMVLLLYS FSDACEEPPT FEAMELIGKP KPYYEIGERV DYKCKKGYFY IPPLATHTIC DRNHTWLPV SDDACYRETC PYIRDPLNGQ AVPANGTYEF GYQMHFICNE GYYLIGEEIL YCELKGSVAI WSGKPPICEK V LCTPPPKI KNGKHTFSEV EVFEYLDAVT YSCDPAPGPD PFSLIGESTI YCGDNSVWSR AAPECKVVKC RFPVVENGKQ IS GFGKKFY YKATVMFECD KGFYLDGSDT IVCDSNSTWD PPVPKCLKVL PPSSTKPPAL SHSVSTSSTT KSPASSASGP RPT YKPPVS NYPGYPKPEE GILDSLDVWV IAVIVIAIVV GVAVICVVPY RYLQRRKKKG TYLTDETHRE VKFTSL UniProtKB: Membrane cofactor protein |
-Macromolecule #3: Desmoglein-2
| Macromolecule | Name: Desmoglein-2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 122.421633 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARSPGRAYA LLLLLICFNV GSGLHLQVLS TRNENKLLPK HPHLVRQKRA WITAPVALRE GEDLSKKNPI AKIHSDLAEE RGLKITYKY TGKGITEPPF GIFVFNKDTG ELNVTSILDR EETPFFLLTG YALDARGNNV EKPLELRIKV LDINDNEPVF T QDVFVGSV ...String: MARSPGRAYA LLLLLICFNV GSGLHLQVLS TRNENKLLPK HPHLVRQKRA WITAPVALRE GEDLSKKNPI AKIHSDLAEE RGLKITYKY TGKGITEPPF GIFVFNKDTG ELNVTSILDR EETPFFLLTG YALDARGNNV EKPLELRIKV LDINDNEPVF T QDVFVGSV EELSAAHTLV MKINATDADE PNTLNSKISY RIVSLEPAYP PVFYLNKDTG EIYTTSVTLD REEHSSYTLT VE ARDGNGE VTDKPVKQAQ VQIRILDVND NIPVVENKVL EGMVEENQVN VEVTRIKVFD ADEIGSDNWL ANFTFASGNE GGY FHIETD AQTNEGIVTL IKEVDYEEMK NLDFSVIVAN KAAFHKSIRS KYKPTPIPIK VKVKNVKEGI HFKSSVISIY VSES MDRSS KGQIIGNFQA FDEDTGLPAH ARYVKLEDRD NWISVDSVTS EIKLAKLPDF ESRYVQNGTY TVKIVAISED YPRKT ITGT VLINVEDIND NCPTLIEPVQ TICHDAEYVN VTAEDLDGHP NSGPFSFSVI DKPPGMAEKW KIARQESTSV LLQQSE KKL GRSEIQFLIS DNQGFSCPEK QVLTLTVCEC LHGSGCREAQ HDSYVGLGPA AIALMILAFL LLLLVPLLLL MCHCGKG AK GFTPIPGTIE MLHPWNNEGA PPEDKVVPSF LPVDQGGSLV GRNGVGGMAK EATMKGSSSA SIVKGQHEMS EMDGRWEE H RSLLSGRATQ FTGATGAIMT TETTKTARAT GASRDMAGAQ AAAVALNEEF LRNYFTDKAA SYTEEDENHT AKDCLLVYS QEETESLNAS IGCCSFIEGE LDDRFLDDLG LKFKTLAEVC LGQKIDINKE IEQRQKPATE TSMNTASHSL CEQTMVNSEN TYSSGSSFP VPKSLQEANA EKVTQEIVTE RSVSSRQAQK VATPLPDPMA SRNVIATETS YVTGSTMPPT TVILGPSQPQ S LIVTERVY APASTLVDQP YANEGTVVVT ERVIQPHGGG SNPLEGTQHL QDVPYVMVRE RESFLAPSSG VQPTLAMPNI AV GQNVTVT ERVLAPASTL QSSYQIPTEN SMTARNTTVS GAGVPGPLPD FGLEESGHSN STITTSSTRV TKHSTVQHSY S UniProtKB: Desmoglein-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X