[English] 日本語
 Yorodumi
Yorodumi- EMDB-18436: Cryo-EM Structure of Human Serine Hydroxymethyltransferase, isofo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Human Serine Hydroxymethyltransferase, isoform 2 (SHMT2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | one-carbon metabolism / folate cycle / tetrahydtofolate /  mitochondria / mitochondria /  TRANSFERASE TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationBRISC complex /  L-allo-threonine aldolase activity / regulation of mitochondrial translation / purine nucleobase biosynthetic process / L-allo-threonine aldolase activity / regulation of mitochondrial translation / purine nucleobase biosynthetic process /  serine binding / L-serine catabolic process / L-serine metabolic process / glycine metabolic process / L-serine biosynthetic process / serine binding / L-serine catabolic process / L-serine metabolic process / glycine metabolic process / L-serine biosynthetic process /  glycine hydroxymethyltransferase ...BRISC complex / glycine hydroxymethyltransferase ...BRISC complex /  L-allo-threonine aldolase activity / regulation of mitochondrial translation / purine nucleobase biosynthetic process / L-allo-threonine aldolase activity / regulation of mitochondrial translation / purine nucleobase biosynthetic process /  serine binding / L-serine catabolic process / L-serine metabolic process / glycine metabolic process / L-serine biosynthetic process / serine binding / L-serine catabolic process / L-serine metabolic process / glycine metabolic process / L-serine biosynthetic process /  glycine hydroxymethyltransferase / glycine hydroxymethyltransferase /  glycine hydroxymethyltransferase activity / glycine biosynthetic process from serine / Metabolism of folate and pterines / glycine hydroxymethyltransferase activity / glycine biosynthetic process from serine / Metabolism of folate and pterines /  regulation of oxidative phosphorylation / tetrahydrofolate metabolic process / response to type I interferon / protein K63-linked deubiquitination / tetrahydrofolate interconversion / regulation of oxidative phosphorylation / tetrahydrofolate metabolic process / response to type I interferon / protein K63-linked deubiquitination / tetrahydrofolate interconversion /  regulation of aerobic respiration / folic acid metabolic process / mitochondrial nucleoid / RHOG GTPase cycle / mRNA regulatory element binding translation repressor activity / protein tetramerization / mRNA 5'-UTR binding / microtubule cytoskeleton / regulation of aerobic respiration / folic acid metabolic process / mitochondrial nucleoid / RHOG GTPase cycle / mRNA regulatory element binding translation repressor activity / protein tetramerization / mRNA 5'-UTR binding / microtubule cytoskeleton /  pyridoxal phosphate binding / one-carbon metabolic process / protein homotetramerization / pyridoxal phosphate binding / one-carbon metabolic process / protein homotetramerization /  mitochondrial inner membrane / mitochondrial inner membrane /  mitochondrial matrix / mitochondrial matrix /  chromatin binding / positive regulation of cell population proliferation / protein homodimerization activity / chromatin binding / positive regulation of cell population proliferation / protein homodimerization activity /  mitochondrion / extracellular exosome / zinc ion binding / mitochondrion / extracellular exosome / zinc ion binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
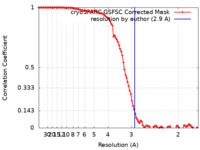
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Rutkiewicz M / Tran LH / Ruszkowski M | |||||||||
| Funding support |  Poland, 1 items Poland, 1 items
| |||||||||
 Citation Citation | Journal: Acta Crystallogr D Struct Biol / Year: 2019 Title: Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Authors: Dorothee Liebschner / Pavel V Afonine / Matthew L Baker / Gábor Bunkóczi / Vincent B Chen / Tristan I Croll / Bradley Hintze / Li Wei Hung / Swati Jain / Airlie J McCoy / Nigel W Moriarty ...Authors: Dorothee Liebschner / Pavel V Afonine / Matthew L Baker / Gábor Bunkóczi / Vincent B Chen / Tristan I Croll / Bradley Hintze / Li Wei Hung / Swati Jain / Airlie J McCoy / Nigel W Moriarty / Robert D Oeffner / Billy K Poon / Michael G Prisant / Randy J Read / Jane S Richardson / David C Richardson / Massimo D Sammito / Oleg V Sobolev / Duncan H Stockwell / Thomas C Terwilliger / Alexandre G Urzhumtsev / Lizbeth L Videau / Christopher J Williams / Paul D Adams /    Abstract: Diffraction (X-ray, neutron and electron) and electron cryo-microscopy are powerful methods to determine three-dimensional macromolecular structures, which are required to understand biological ...Diffraction (X-ray, neutron and electron) and electron cryo-microscopy are powerful methods to determine three-dimensional macromolecular structures, which are required to understand biological processes and to develop new therapeutics against diseases. The overall structure-solution workflow is similar for these techniques, but nuances exist because the properties of the reduced experimental data are different. Software tools for structure determination should therefore be tailored for each method. Phenix is a comprehensive software package for macromolecular structure determination that handles data from any of these techniques. Tasks performed with Phenix include data-quality assessment, map improvement, model building, the validation/rebuilding/refinement cycle and deposition. Each tool caters to the type of experimental data. The design of Phenix emphasizes the automation of procedures, where possible, to minimize repetitive and time-consuming manual tasks, while default parameters are chosen to encourage best practice. A graphical user interface provides access to many command-line features of Phenix and streamlines the transition between programs, project tracking and re-running of previous tasks. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18436.map.gz emd_18436.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18436-v30.xml emd-18436-v30.xml emd-18436.xml emd-18436.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18436_fsc.xml emd_18436_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18436.png emd_18436.png | 145 KB | ||
| Filedesc metadata |  emd-18436.cif.gz emd-18436.cif.gz | 6.8 KB | ||
| Others |  emd_18436_half_map_1.map.gz emd_18436_half_map_1.map.gz emd_18436_half_map_2.map.gz emd_18436_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18436 http://ftp.pdbj.org/pub/emdb/structures/EMD-18436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18436 | HTTPS FTP |
-Related structure data
| Related structure data |  8qi7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18436.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18436.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18436_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
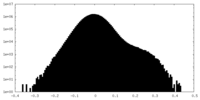
| Projections & Slices |
| ||||||||||||
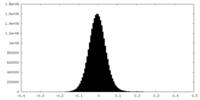
| Density Histograms |
-Half map: #2
| File | emd_18436_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
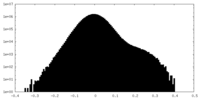
| Projections & Slices |
| ||||||||||||
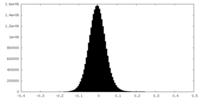
| Density Histograms |
- Sample components
Sample components
-Entire : SHMT2 in the form of PLP internal aldimine
| Entire | Name: SHMT2 in the form of PLP internal aldimine Serine hydroxymethyltransferase Serine hydroxymethyltransferase |
|---|---|
| Components |
|
-Supramolecule #1: SHMT2 in the form of PLP internal aldimine
| Supramolecule | Name: SHMT2 in the form of PLP internal aldimine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Serine hydroxymethyltransferase, mitochondrial
| Macromolecule | Name: Serine hydroxymethyltransferase, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number:  glycine hydroxymethyltransferase glycine hydroxymethyltransferase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.944973 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: SNAAQTQTGE ANRGWTGQES LSDSDPEMWE LLQREKDRQC RGLELIASEN FCSRAALEAL GSCLNNKYSE GYPGKRYYGG AEVVDEIEL LCQRRALEAF DLDPAQWGVN VQPYSGSPAN LAVYTALLQP HDRIMGLDLP DGGHLTHGYM SDVKRISATS I FFESMPYK ...String: SNAAQTQTGE ANRGWTGQES LSDSDPEMWE LLQREKDRQC RGLELIASEN FCSRAALEAL GSCLNNKYSE GYPGKRYYGG AEVVDEIEL LCQRRALEAF DLDPAQWGVN VQPYSGSPAN LAVYTALLQP HDRIMGLDLP DGGHLTHGYM SDVKRISATS I FFESMPYK LNPKTGLIDY NQLALTARLF RPRLIIAGTS AYARLIDYAR MREVCDEVKA HLLADMAHIS GLVAAKVIPS PF KHADIVT TTTH(LLP)TLRGA RSGLIFYRKG VKAVDPKTGR EIPYTFEDRI NFAVFPSLQG GPHNHAIAAV AVALKQACT PMFREYSLQV LKNARAMADA LLERGYSLVS GGTDNHLVLV DLRPKGLDGA RAERVLELVS ITANKNTCPG DRSAITPGGL RLGAPALTS RQFREDDFRR VVDFIDEGVN IGLEVKSKTA KLQDFKSFLL KDSETSQRLA NLRQRVEQFA RAFPMPGFDE H UniProtKB:  Serine hydroxymethyltransferase, mitochondrial Serine hydroxymethyltransferase, mitochondrial |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 117 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25 mM Hepes pH 7.5, 150 mM NaCl, 1 mM TCEP |
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 7948 / Average electron dose: 40.4 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X