[English] 日本語
 Yorodumi
Yorodumi- EMDB-18325: Respiratory complex I from Paracoccus denitrificans in MSP2N2 nan... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Respiratory complex I from Paracoccus denitrificans in MSP2N2 nanodiscs (ND4 & ND5 focus refinement) | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Respiratory complex I / NADH:ubiquinone oxidoreductase / Nanodiscs / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationNADH:ubiquinone reductase (H+-translocating) / NADH dehydrogenase (ubiquinone) activity / ATP synthesis coupled electron transport / membrane Similarity search - Function | |||||||||
| Biological species |  Paracoccus denitrificans PD1222 (bacteria) Paracoccus denitrificans PD1222 (bacteria) | |||||||||
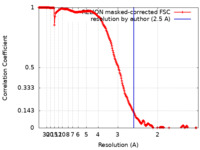
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Ivanov BS / Bridges HR / Hirst J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Respiratory complex I from Paracoccus denitrificans Authors: Ivanov BS / Bridges HR / Hirst J / Jarman OD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18325.map.gz emd_18325.map.gz | 917.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18325-v30.xml emd-18325-v30.xml emd-18325.xml emd-18325.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18325_fsc.xml emd_18325_fsc.xml | 22.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18325.png emd_18325.png | 147.1 KB | ||
| Masks |  emd_18325_msk_1.map emd_18325_msk_1.map | 1000 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18325.cif.gz emd-18325.cif.gz | 6.6 KB | ||
| Others |  emd_18325_half_map_1.map.gz emd_18325_half_map_1.map.gz emd_18325_half_map_2.map.gz emd_18325_half_map_2.map.gz | 811.3 MB 811.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18325 http://ftp.pdbj.org/pub/emdb/structures/EMD-18325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18325 | HTTPS FTP |
-Validation report
| Summary document |  emd_18325_validation.pdf.gz emd_18325_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18325_full_validation.pdf.gz emd_18325_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_18325_validation.xml.gz emd_18325_validation.xml.gz | 30.7 KB | Display | |
| Data in CIF |  emd_18325_validation.cif.gz emd_18325_validation.cif.gz | 40.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18325 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18325 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18325 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18325 | HTTPS FTP |
-Related structure data
| Related structure data |  8qc1MC  8qbyC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18325.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18325.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.745 Å | ||||||||||||||||||||||||||||||||||||
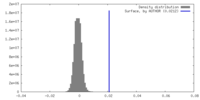
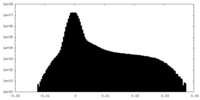
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18325_msk_1.map emd_18325_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
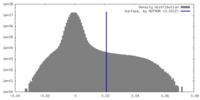
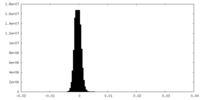
| Density Histograms |
-Half map: half-map 1
| File | emd_18325_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
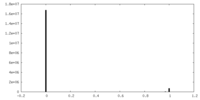
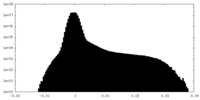
| Density Histograms |
-Half map: half-map 2
| File | emd_18325_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
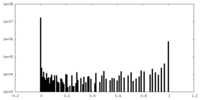
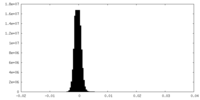
| Density Histograms |
- Sample components
Sample components
-Entire : Respiratory complex I
| Entire | Name: Respiratory complex I |
|---|---|
| Components |
|
-Supramolecule #1: Respiratory complex I
| Supramolecule | Name: Respiratory complex I / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Paracoccus denitrificans PD1222 (bacteria) Paracoccus denitrificans PD1222 (bacteria) |
-Macromolecule #1: NADH dehydrogenase subunit M
| Macromolecule | Name: NADH dehydrogenase subunit M / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH:ubiquinone reductase (H+-translocating) |
|---|---|
| Source (natural) | Organism:  Paracoccus denitrificans PD1222 (bacteria) Paracoccus denitrificans PD1222 (bacteria) |
| Molecular weight | Theoretical: 56.519906 KDa |
| Sequence | String: MTNLLSIITF LPIVAAIIMA LFLRGQDEAA ARNAKWLALL TTTATFVISL FVLFRFDPAN TGFQFVEDHA WIMGLRYKMG VDGISVLFV LLTTFMMPLT ILSTWQVQDK VKEYMIAFLV LEGLMIGVFT ALDLVLFYLF FEAGLIPMFL IIGIWGGKDR I YASFKFFL ...String: MTNLLSIITF LPIVAAIIMA LFLRGQDEAA ARNAKWLALL TTTATFVISL FVLFRFDPAN TGFQFVEDHA WIMGLRYKMG VDGISVLFV LLTTFMMPLT ILSTWQVQDK VKEYMIAFLV LEGLMIGVFT ALDLVLFYLF FEAGLIPMFL IIGIWGGKDR I YASFKFFL YTFLGSVLML VAMIAMYRMA GTTDIPTLLT FDFPSENFRL LGMTVVGGMQ MLLFLAFFAS FAVKMPMWPV HT WLPDAHV QAPTAGSVLL AAVLLKMGGY GFLRFSLPMF PVASGVAQPY VFWLSAIAIV YTSLVALAQS DMKKVIAYSS VAH MGYVTM GVFAANQIGV DGAIFQMLSH GFISGALFLC VGVIYDRMHT REIDAYGGLV NRMPAYAAVF MFFTMANVGL PGTS GFVGE FLTLMGVFRV DTWVALVATS GVILSAAYAL WLYRRVTLGQ LIKESLKSIT DMTPRERWVF IPLIAMTLIL GVYPR LVTD VTGPAVAALV QDYNQSQPAA PVATAQASH UniProtKB: NADH dehydrogenase subunit M |
-Macromolecule #2: NADH dehydrogenase subunit L
| Macromolecule | Name: NADH dehydrogenase subunit L / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH:ubiquinone reductase (H+-translocating) |
|---|---|
| Source (natural) | Organism:  Paracoccus denitrificans PD1222 (bacteria) Paracoccus denitrificans PD1222 (bacteria) |
| Molecular weight | Theoretical: 77.811352 KDa |
| Sequence | String: MEKFVLFAPL IASLIAGLGW RAIGEKAAQY LTTGVLFLSC LISWYLFLSF DGVPRHIPVL DWVVTGDFHA EWAIRLDRLT AIMLIVVTT VSALVHMYSL GYMAHDDNWT HDEHYKARFF AYLSFFTFAM LMLVTADNLL QMFFGWEGVG VASYLLIGFY Y KKASANAA ...String: MEKFVLFAPL IASLIAGLGW RAIGEKAAQY LTTGVLFLSC LISWYLFLSF DGVPRHIPVL DWVVTGDFHA EWAIRLDRLT AIMLIVVTT VSALVHMYSL GYMAHDDNWT HDEHYKARFF AYLSFFTFAM LMLVTADNLL QMFFGWEGVG VASYLLIGFY Y KKASANAA AMKAFIVNRV GDFGFLLGIF GIYWLTGSVQ FDEIFRQVPQ LAQTEMHFLW RDWNAANLLG FLLFVGAMGK SA QLLLHTW LPDAMEGPTP VSALIHAATM VTAGVFLVCR MSPLYEFAPD AKNFIVIIGA TTAFFAATVG LVQNDIKRVI AYS TCSQLG YMFVAAGVGV YSAAMFHLLT HAFFKAMLFL GAGSVIHAMH HEQDMRNYGG LRKKIPLTFW AMMIGTFAIT GVGI PLTHL GFAGFLSKDA IIESAYAGSG YAFWLLVIAA CFTSFYSWRL IFLTFYGKPR GDHHAHDHAH ESPPVMTIPL GVLAI GAVF AGMVWYGPFF GDHHKVTEYF HIAGAHHEAA EGEEAEHATA EAPVEHAVAD TATAEGEAAA EAEHAEIAAP VGGAIY MHP DNHIMDEAHH APAWVKVSPF VAMVLGLITA WTFYIANPSL PRRLAAQQPA LYRFLLNKWY FDEIYEFIFV RPAKWLG RV LWKGGDGAVI DGTINGVAMG LIPRLTRAAV RVQSGYLFHY AFAMVLGIVG LLIWVMMRGA H UniProtKB: NADH dehydrogenase subunit L |
-Macromolecule #3: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
| Macromolecule | Name: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE / type: ligand / ID: 3 / Number of copies: 8 / Formula: 3PH |
|---|---|
| Molecular weight | Theoretical: 704.998 Da |
| Chemical component information |  ChemComp-3PH: |
-Macromolecule #4: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 4 / Number of copies: 4 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #5: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 5 / Number of copies: 1 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #7: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phospho...
| Macromolecule | Name: O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine type: ligand / ID: 7 / Number of copies: 1 / Formula: P5S |
|---|---|
| Molecular weight | Theoretical: 792.075 Da |
| Chemical component information |  ChemComp-P5S: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 115 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 3 / Number real images: 16814 / Average exposure time: 2.4 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model / Details: Model Angelo |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8qc1: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)