[English] 日本語
 Yorodumi
Yorodumi- EMDB-18127: S-layer of archaeon Sulfolobus acidocaldarius by subtomogram averaging -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S-layer of archaeon Sulfolobus acidocaldarius by subtomogram averaging | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | S-layer / protein-protein interactions / protein-membrane interactions / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Sulfolobus acidocaldarius (acidophilic) Sulfolobus acidocaldarius (acidophilic) | |||||||||
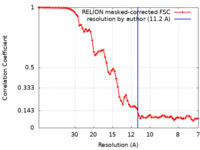
| Method | subtomogram averaging / cryo EM / Resolution: 11.2 Å | |||||||||
 Authors Authors | Gambelli L / McLaren MJ / Daum B | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structure of the two-component S-layer of the archaeon . Authors: Lavinia Gambelli / Mathew McLaren / Rebecca Conners / Kelly Sanders / Matthew C Gaines / Lewis Clark / Vicki A M Gold / Daniel Kattnig / Mateusz Sikora / Cyril Hanus / Michail N Isupov / Bertram Daum /     Abstract: Surface layers (S-layers) are resilient two-dimensional protein lattices that encapsulate many bacteria and most archaea. In archaea, S-layers usually form the only structural component of the cell ...Surface layers (S-layers) are resilient two-dimensional protein lattices that encapsulate many bacteria and most archaea. In archaea, S-layers usually form the only structural component of the cell wall and thus act as the final frontier between the cell and its environment. Therefore, S-layers are crucial for supporting microbial life. Notwithstanding their importance, little is known about archaeal S-layers at the atomic level. Here, we combined single-particle cryo electron microscopy, cryo electron tomography, and Alphafold2 predictions to generate an atomic model of the two-component S-layer of . The outer component of this S-layer (SlaA) is a flexible, highly glycosylated, and stable protein. Together with the inner and membrane-bound component (SlaB), they assemble into a porous and interwoven lattice. We hypothesise that jackknife-like conformational changes in SlaA play important roles in S-layer assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18127.map.gz emd_18127.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18127-v30.xml emd-18127-v30.xml emd-18127.xml emd-18127.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18127_fsc.xml emd_18127_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_18127.png emd_18127.png | 56.9 KB | ||
| Filedesc metadata |  emd-18127.cif.gz emd-18127.cif.gz | 5.8 KB | ||
| Others |  emd_18127_half_map_1.map.gz emd_18127_half_map_1.map.gz emd_18127_half_map_2.map.gz emd_18127_half_map_2.map.gz | 15.5 MB 15.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18127 http://ftp.pdbj.org/pub/emdb/structures/EMD-18127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18127 | HTTPS FTP |
-Validation report
| Summary document |  emd_18127_validation.pdf.gz emd_18127_validation.pdf.gz | 920.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18127_full_validation.pdf.gz emd_18127_full_validation.pdf.gz | 920.4 KB | Display | |
| Data in XML |  emd_18127_validation.xml.gz emd_18127_validation.xml.gz | 11.7 KB | Display | |
| Data in CIF |  emd_18127_validation.cif.gz emd_18127_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18127 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18127 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18127 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18127 | HTTPS FTP |
-Related structure data
| Related structure data |  8qoxMC  8qp0MC  7zcxC  8an2C  8an3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18127.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18127.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18127_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
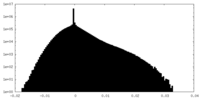
| Projections & Slices |
| ||||||||||||
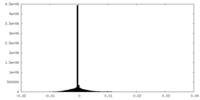
| Density Histograms |
-Half map: #1
| File | emd_18127_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
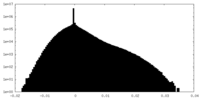
| Projections & Slices |
| ||||||||||||
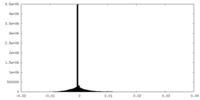
| Density Histograms |
- Sample components
Sample components
-Entire : Exosome
| Entire | Name: Exosome |
|---|---|
| Components |
|
-Supramolecule #1: Exosome
| Supramolecule | Name: Exosome / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Sulfolobus acidocaldarius (acidophilic) / Strain: MW001 Sulfolobus acidocaldarius (acidophilic) / Strain: MW001 |
-Macromolecule #1: SlaA S-layer
| Macromolecule | Name: SlaA S-layer / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Sulfolobus acidocaldarius (acidophilic) / Strain: MW001 Sulfolobus acidocaldarius (acidophilic) / Strain: MW001 |
| Sequence | String: MNKLVGLLVS SLFLASILIG IAPAITTTAL TPPVSAGGIQ AYLLTGSGAP ASGLVLFVVN VSNIQVSSS NVTNVISTVV SNIQINAKTE NAQTGATTGS VTVRFPTSGY NAYYDSVDKV V FVVVSFLY PYTTTSVNIP LSYLSKYLPG LLTAQPYDET GAQVTSVSST ...String: MNKLVGLLVS SLFLASILIG IAPAITTTAL TPPVSAGGIQ AYLLTGSGAP ASGLVLFVVN VSNIQVSSS NVTNVISTVV SNIQINAKTE NAQTGATTGS VTVRFPTSGY NAYYDSVDKV V FVVVSFLY PYTTTSVNIP LSYLSKYLPG LLTAQPYDET GAQVTSVSST PFGSLIDTST GQ QILGTNP VLTSYNSYTT QANTNMQEGV VSGTLTSFTL GGQSFSGSTV PVILYAPFIF SNS PYQAGL YNPMQVNGNL GSLSSEAYYH PVIWGRALIN TTLIDTYASG SVPFTFQLNY SVPG PLTIN MAQLAWIASI NNLPTSFTYL SYKFSNGYES FLGIISNSTQ LTAGALTINP SGNFT INGK KFYVYLLVVG STNSTTPVEY VTKLVVEYPS STNFLPQGVT VTTSSNKYTL PVYEIG GPA GTTITLTGNW YSTPYTVQIT VGSTPTLTNY VSQILLKAVA YEGINVSTTQ SPYYSTA IL STPPSEISIT GSSTITAQGK LTATSASATV NLLTNATLTY ENIPLTQYSF NGIIVTPG Y AAINGTTAMA YVIGALYNKT SDYVLSFAGS QEPMQVMNNN LTEVTTLAPF GLTLLAPSV PATETGTSPL QLEFFTVPST SYIALVDFGL WGNLTSVTVS AYDTVNNKLS VNLGYFYGIV IPPSISTAP YNYQNFICPN NYVTVTIYDP DAVLDPYPSG SFTTSSLPLK YGNMNITGAV I FPGSSVYN PSGVFGYSNF NKGAAVTTFT YTAQSGPFSP VALTGNTNYL SQYADNNPTD NY YFIQTVN GMPVLMGGLS IVASPVSASL PSSTSSPGFM YLLPSAAQVP SPLPGMATPN YNL NIYITY KIDGATVGNN MINGLYVASQ NTLIYVVPNG SFVGSNIKLT YTTTDYAVLH YFYS TGQYK VFKTVSVPNV TANLYFPSST TPLYQLSVPL YLSEPYYGSP LPTYIGLGTN GTSLW NSPN YVLFGVSAVQ QYLGFIKSIS VTLSNGTTVV IPLTTSNMQT LFPQLVGQEL QACNGT FQF GISITGLEKL LNLNVQQLNN SILSVTYHDY VTGETLTATT KLVALSTLSL VAKGAGV VE FLLTAYPYTG NITFAPPWFI AENVVKQPFM TYSDLQFAKT NPSAILSLST VNITVVGL G GKASVYYNST SGQTVITNIY GQTVATLSGN VLPTLTELAA GNGTFTGSLQ FTIVPNNTV VQIPSSLTKT SFAVYTNGSL AIVLNGKAYS LGPAGLFLLP FVTYTGSAIG ANATAIITVS DGVGTSTTQ VPITAENFTP IRLAPFQVPA QVPLPNAPKL KYEYNGSIVI TPQQQVLKIY V TSILPYPQ EFQIQAFVYE ASQFNVHTGS PTAAPVYFSY SAVRAYPALG IGTSVPNLLV YV QLQGISN LPAGKYVIVL SAVPFAGGPV LSEYPAQLIF TNVTLTQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | TFS KRIOS |
| Image recording | Image recording ID: 1 / Film or detector model: TFS FALCON 4i (4k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average electron dose: 2.02 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | TFS TALOS |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 2.02 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)