[English] 日本語
 Yorodumi
Yorodumi- EMDB-17741: Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleosome / nuclear receptor / NR5A2 / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of chromatin / nucleosome / protein heterodimerization activity / DNA binding / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
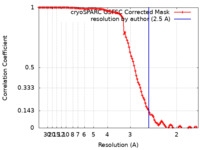
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Kobayashi W / Sappler A / Bollschweiler D / Kummecke M / Basquin J / Arslantas E / Ruangroengkulrith S / Hornberger R / Duderstadt K / Tachibana K | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition. Authors: Wataru Kobayashi / Anna H Sappler / Daniel Bollschweiler / Maximilian Kümmecke / Jérôme Basquin / Eda Nur Arslantas / Siwat Ruangroengkulrith / Renate Hornberger / Karl Duderstadt / Kikuë Tachibana /  Abstract: Gene expression during natural and induced reprogramming is controlled by pioneer transcription factors that initiate transcription from closed chromatin. Nr5a2 is a key pioneer factor that regulates ...Gene expression during natural and induced reprogramming is controlled by pioneer transcription factors that initiate transcription from closed chromatin. Nr5a2 is a key pioneer factor that regulates zygotic genome activation in totipotent embryos, pluripotency in embryonic stem cells and metabolism in adult tissues, but the mechanism of its pioneer activity remains poorly understood. Here, we present a cryo-electron microscopy structure of human NR5A2 bound to a nucleosome. The structure shows that the conserved carboxy-terminal extension (CTE) loop of the NR5A2 DNA-binding domain competes with a DNA minor groove anchor of the nucleosome and releases entry-exit site DNA. Mutational analysis showed that NR5A2 D159 of the CTE is dispensable for DNA binding but required for stable nucleosome association and persistent DNA 'unwrapping'. These findings suggest that NR5A2 belongs to an emerging class of pioneer factors that can use DNA minor groove anchor competition to destabilize nucleosomes and facilitate gene expression during reprogramming. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17741.map.gz emd_17741.map.gz | 63 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17741-v30.xml emd-17741-v30.xml emd-17741.xml emd-17741.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17741_fsc.xml emd_17741_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17741.png emd_17741.png | 64.3 KB | ||
| Filedesc metadata |  emd-17741.cif.gz emd-17741.cif.gz | 6.2 KB | ||
| Others |  emd_17741_additional_1.map.gz emd_17741_additional_1.map.gz emd_17741_half_map_1.map.gz emd_17741_half_map_1.map.gz emd_17741_half_map_2.map.gz emd_17741_half_map_2.map.gz | 14 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17741 http://ftp.pdbj.org/pub/emdb/structures/EMD-17741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17741 | HTTPS FTP |
-Validation report
| Summary document |  emd_17741_validation.pdf.gz emd_17741_validation.pdf.gz | 870.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17741_full_validation.pdf.gz emd_17741_full_validation.pdf.gz | 870 KB | Display | |
| Data in XML |  emd_17741_validation.xml.gz emd_17741_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_17741_validation.cif.gz emd_17741_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17741 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17741 | HTTPS FTP |
-Related structure data
| Related structure data |  8pkjMC  8pkiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17741.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17741.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||
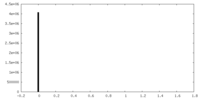
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: lower resolution map where the end of the DNA is better define
| File | emd_17741_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | lower resolution map where the end of the DNA is better define | ||||||||||||
| Projections & Slices |
| ||||||||||||
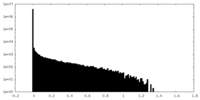
| Density Histograms |
-Half map: #2
| File | emd_17741_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
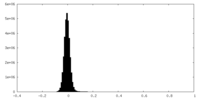
| Density Histograms |
-Half map: #1
| File | emd_17741_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
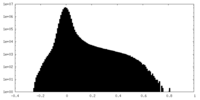
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5
| Entire | Name: Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5
| Supramolecule | Name: Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Histone H3 (Fragment)
| Macromolecule | Name: Histone H3 (Fragment) / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.360983 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PSTGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSAA IGALQEASEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.394426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.165551 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKQGGK ARAKAKTRSS RAGLQFPVGR VHRLLRKGNY SERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG RVTIAQGGVL PNIQAVLLPK KTESHHKAKG K UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.937213 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPEPAKSAPA PKKGSKKAVT KAQKKDGKKR KRSRKESYSV YVYKVLKQVH PDTGISSKAM GIMNSFVNDI FERIAGEASR LAHYNKRST ITSREIQTAV RLLLPGELAK HAVSEGTKAV TKYTSSK UniProtKB: Histone H2B |
-Macromolecule #5: DNA
| Macromolecule | Name: DNA / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 46.968922 KDa |
| Sequence | String: (DA)(DT)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DA)(DT)(DC)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG) (DT)(DA)(DG)(DA)(DC)(DA) ...String: (DA)(DT)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DA)(DT)(DC)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG) (DT)(DA)(DG)(DA)(DC)(DA)(DG)(DC) (DT)(DC)(DT)(DA)(DG)(DC)(DA)(DC)(DC)(DG) (DC)(DT) (DT)(DA)(DA)(DA)(DC)(DG)(DC) (DA)(DC)(DG)(DT)(DA)(DC)(DG)(DC)(DG)(DC) (DT)(DG)(DT) (DC)(DC)(DC)(DC)(DC)(DG) (DC)(DG)(DT)(DT)(DT)(DT)(DA)(DA)(DC)(DC) (DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG)(DG) (DA)(DT)(DT)(DA)(DC)(DT)(DC)(DC)(DC)(DT) (DA)(DG)(DT)(DC)(DT) (DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC)(DG)(DT)(DT)(DC)(DA)(DA) (DG)(DG)(DC)(DC)(DA)(DA) (DT)(DA)(DC) (DA)(DT)(DC)(DC)(DT)(DG)(DT)(DG)(DA)(DT) |
-Macromolecule #6: DNA
| Macromolecule | Name: DNA / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 47.489234 KDa |
| Sequence | String: (DA)(DT)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DT)(DG)(DG)(DC)(DC)(DT) (DT)(DG)(DA)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA) ...String: (DA)(DT)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DT)(DG)(DG)(DC)(DC)(DT) (DT)(DG)(DA)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA)(DA)(DT) (DC)(DC)(DC)(DC)(DT)(DT)(DG)(DG)(DC)(DG) (DG)(DT) (DT)(DA)(DA)(DA)(DA)(DC)(DG) (DC)(DG)(DG)(DG)(DG)(DG)(DA)(DC)(DA)(DG) (DC)(DG)(DC) (DG)(DT)(DA)(DC)(DG)(DT) (DG)(DC)(DG)(DT)(DT)(DT)(DA)(DA)(DG)(DC) (DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG)(DA) (DG)(DC)(DT)(DG)(DT)(DC)(DT)(DA)(DC)(DG) (DA)(DC)(DC)(DA)(DA) (DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG)(DC)(DC)(DT)(DC)(DG)(DG) (DC)(DA)(DC)(DC)(DG)(DG) (DG)(DA)(DT) (DT)(DC)(DT)(DC)(DC)(DA)(DG)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)