[English] 日本語
 Yorodumi
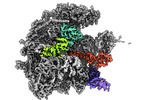
Yorodumi- EMDB-17721: human mitoribosomal large subunit assembly intermediate 3 with GTPBP5 -
+ Open data
Open data
- Basic information
Basic information
| Entry | 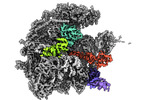 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human mitoribosomal large subunit assembly intermediate 3 with GTPBP5 | |||||||||
 Map data Map data | full map, sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondria / maturation / biogenesis / ribosome / GTPBP5 / OBGH1 / LSU | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Kummer E / Nguyen TG | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural insights into the role of GTPBP10 in the RNA maturation of the mitoribosome. Authors: Thu Giang Nguyen / Christina Ritter / Eva Kummer /  Abstract: Mitochondria contain their own genetic information and a dedicated translation system to express it. The mitochondrial ribosome is assembled from mitochondrial-encoded RNA and nuclear-encoded ...Mitochondria contain their own genetic information and a dedicated translation system to express it. The mitochondrial ribosome is assembled from mitochondrial-encoded RNA and nuclear-encoded ribosomal proteins. Assembly is coordinated in the mitochondrial matrix by biogenesis factors that transiently associate with the maturing particle. Here, we present a structural snapshot of a large mitoribosomal subunit assembly intermediate containing 7 biogenesis factors including the GTPases GTPBP7 and GTPBP10. Our structure illustrates how GTPBP10 aids the folding of the ribosomal RNA during the biogenesis process, how this process is related to bacterial ribosome biogenesis, and why mitochondria require two biogenesis factors in contrast to only one in bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17721.map.gz emd_17721.map.gz | 398.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17721-v30.xml emd-17721-v30.xml emd-17721.xml emd-17721.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17721.png emd_17721.png | 117.6 KB | ||
| Masks |  emd_17721_msk_1.map emd_17721_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17721.cif.gz emd-17721.cif.gz | 4 KB | ||
| Others |  emd_17721_additional_1.map.gz emd_17721_additional_1.map.gz emd_17721_half_map_1.map.gz emd_17721_half_map_1.map.gz emd_17721_half_map_2.map.gz emd_17721_half_map_2.map.gz | 212.3 MB 391 MB 390.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17721 http://ftp.pdbj.org/pub/emdb/structures/EMD-17721 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17721 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17721 | HTTPS FTP |
-Validation report
| Summary document |  emd_17721_validation.pdf.gz emd_17721_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17721_full_validation.pdf.gz emd_17721_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_17721_validation.xml.gz emd_17721_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_17721_validation.cif.gz emd_17721_validation.cif.gz | 21.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17721 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17721 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17721 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17721 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17721.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17721.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map, sharpened | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17721_msk_1.map emd_17721_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
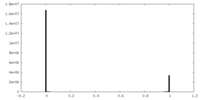
| Density Histograms |
-Additional map: full map, unsharpened
| File | emd_17721_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map, unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
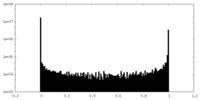
| Density Histograms |
-Half map: #1
| File | emd_17721_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
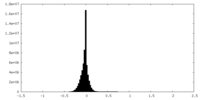
| Density Histograms |
-Half map: #2
| File | emd_17721_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
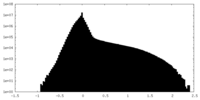
| Density Histograms |
- Sample components
Sample components
-Entire : large mitoribosomal subunit biogenesis intermediate with GTPBP10,...
| Entire | Name: large mitoribosomal subunit biogenesis intermediate with GTPBP10, GTPBP7, NSUN4-MTERF4, ACP-module |
|---|---|
| Components |
|
-Supramolecule #1: large mitoribosomal subunit biogenesis intermediate with GTPBP10,...
| Supramolecule | Name: large mitoribosomal subunit biogenesis intermediate with GTPBP10, GTPBP7, NSUN4-MTERF4, ACP-module type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#58 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.9 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.05 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 39296 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X