+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CAK in complex with inhibitor dinaciclib | |||||||||
 Map data Map data | Post-processed sharpened cryo-EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / Inhibitor / Transcription / Cell Cycle / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationventricular system development / snRNA transcription by RNA polymerase II / CAK-ERCC2 complex / transcription factor TFIIK complex / adult heart development / transcription factor TFIIH holo complex / transcription factor TFIIH core complex / cyclin-dependent protein serine/threonine kinase activator activity / [RNA-polymerase]-subunit kinase / RNA Polymerase I Transcription Termination ...ventricular system development / snRNA transcription by RNA polymerase II / CAK-ERCC2 complex / transcription factor TFIIK complex / adult heart development / transcription factor TFIIH holo complex / transcription factor TFIIH core complex / cyclin-dependent protein serine/threonine kinase activator activity / [RNA-polymerase]-subunit kinase / RNA Polymerase I Transcription Termination / cyclin-dependent protein serine/threonine kinase regulator activity / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / regulation of G1/S transition of mitotic cell cycle / RNA Polymerase I Transcription Initiation / RNA polymerase II transcribes snRNA genes / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / cyclin-dependent kinase / Formation of HIV elongation complex in the absence of HIV Tat / cyclin-dependent protein serine/threonine kinase activity / ATP-dependent activity, acting on DNA / Cyclin E associated events during G1/S transition / RNA Polymerase II Transcription Elongation / Cyclin A/B1/B2 associated events during G2/M transition / cyclin-dependent protein kinase holoenzyme complex / Formation of RNA Pol II elongation complex / Cyclin A:Cdk2-associated events at S phase entry / RNA Polymerase II Pre-transcription Events / RNA polymerase II CTD heptapeptide repeat kinase activity / male germ cell nucleus / TP53 Regulates Transcription of DNA Repair Genes / nucleotide-excision repair / transcription initiation at RNA polymerase II promoter / RNA Polymerase I Promoter Escape / positive regulation of smooth muscle cell proliferation / NoRC negatively regulates rRNA expression / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / fibrillar center / Formation of Incision Complex in GG-NER / response to calcium ion / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / G1/S transition of mitotic cell cycle / Cyclin D associated events in G1 / RUNX1 regulates transcription of genes involved in differentiation of HSCs / transcription by RNA polymerase II / regulation of cell cycle / protein stabilization / protein kinase activity / cell division / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / negative regulation of apoptotic process / regulation of transcription by RNA polymerase II / perinuclear region of cytoplasm / positive regulation of transcription by RNA polymerase II / zinc ion binding / nucleoplasm / ATP binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.9 Å | |||||||||
 Authors Authors | Cushing VI / Koh AF / Feng J / Jurgaityte K / Bahl AK / Ali S / Kotecha A / Greber BJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design. Authors: Victoria I Cushing / Adrian F Koh / Junjie Feng / Kaste Jurgaityte / Alexander Bondke / Sebastian H B Kroll / Marion Barbazanges / Bodo Scheiper / Ash K Bahl / Anthony G M Barrett / Simak ...Authors: Victoria I Cushing / Adrian F Koh / Junjie Feng / Kaste Jurgaityte / Alexander Bondke / Sebastian H B Kroll / Marion Barbazanges / Bodo Scheiper / Ash K Bahl / Anthony G M Barrett / Simak Ali / Abhay Kotecha / Basil J Greber /     Abstract: Rational design of next-generation therapeutics can be facilitated by high-resolution structures of drug targets bound to small-molecule inhibitors. However, application of structure-based methods to ...Rational design of next-generation therapeutics can be facilitated by high-resolution structures of drug targets bound to small-molecule inhibitors. However, application of structure-based methods to macromolecules refractory to crystallization has been hampered by the often-limiting resolution and throughput of cryogenic electron microscopy (cryo-EM). Here, we use high-resolution cryo-EM to determine structures of the CDK-activating kinase, a master regulator of cell growth and division, in its free and nucleotide-bound states and in complex with 15 inhibitors at up to 1.8 Å resolution. Our structures provide detailed insight into inhibitor interactions and networks of water molecules in the active site of cyclin-dependent kinase 7 and provide insights into the mechanisms contributing to inhibitor selectivity, thereby providing the basis for rational design of next-generation therapeutics. These results establish a methodological framework for the use of high-resolution cryo-EM in structure-based drug design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17521.map.gz emd_17521.map.gz | 202.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17521-v30.xml emd-17521-v30.xml emd-17521.xml emd-17521.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17521.png emd_17521.png | 134.7 KB | ||
| Masks |  emd_17521_msk_1.map emd_17521_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17521.cif.gz emd-17521.cif.gz | 7 KB | ||
| Others |  emd_17521_half_map_1.map.gz emd_17521_half_map_1.map.gz emd_17521_half_map_2.map.gz emd_17521_half_map_2.map.gz | 172.9 MB 172.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17521 http://ftp.pdbj.org/pub/emdb/structures/EMD-17521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17521 | HTTPS FTP |
-Validation report
| Summary document |  emd_17521_validation.pdf.gz emd_17521_validation.pdf.gz | 1010.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17521_full_validation.pdf.gz emd_17521_full_validation.pdf.gz | 1009.7 KB | Display | |
| Data in XML |  emd_17521_validation.xml.gz emd_17521_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_17521_validation.cif.gz emd_17521_validation.cif.gz | 18.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17521 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17521 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17521 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17521 | HTTPS FTP |
-Related structure data
| Related structure data |  8p78MC  8ormC  8p6vC  8p6wC  8p6xC  8p6yC  8p6zC  8p70C  8p71C  8p72C  8p73C  8p74C  8p75C  8p76C  8p77C  8p79C  8p7lC  8plzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17521.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17521.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed sharpened cryo-EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7125 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17521_msk_1.map emd_17521_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half-map
| File | emd_17521_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half-map
| File | emd_17521_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CDK-activating kinase
| Entire | Name: CDK-activating kinase |
|---|---|
| Components |
|
-Supramolecule #1: CDK-activating kinase
| Supramolecule | Name: CDK-activating kinase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: CAK in complex with non-covalent inhibitor dinaciclib |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 85 KDa |
-Macromolecule #1: CDK-activating kinase assembly factor MAT1
| Macromolecule | Name: CDK-activating kinase assembly factor MAT1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.234531 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SNAPVTFSTG IKMGQHISLA PIHKLEEALY EYQPLQIETY GPHVPELEML GRLGYLNHVR AASPQDLAGG YTSSLACHRA LQDAFSGLF WQPS UniProtKB: CDK-activating kinase assembly factor MAT1 |
-Macromolecule #2: Cyclin-H
| Macromolecule | Name: Cyclin-H / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.721508 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: (ACE)MYHNSSQKR HWTFSSEEQL ARLRADANRK FRCKAVANGK VLPNDPVFLE PHEEMTLCKY YEKRLLEFCS VFKPAM PRS VVGTACMYFK RFYLNNSVME YHPRIIMLTC AFLACKVDEF NVSSPQFVGN LRESPLGQEK ALEQILEYEL LLIQQLN FH LIVHNPYRPF ...String: (ACE)MYHNSSQKR HWTFSSEEQL ARLRADANRK FRCKAVANGK VLPNDPVFLE PHEEMTLCKY YEKRLLEFCS VFKPAM PRS VVGTACMYFK RFYLNNSVME YHPRIIMLTC AFLACKVDEF NVSSPQFVGN LRESPLGQEK ALEQILEYEL LLIQQLN FH LIVHNPYRPF EGFLIDLKTR YPILENPEIL RKTADDFLNR IALTDAYLLY TPSQIALTAI LSSASRAGIT MESYLSES L MLKENRTCLS QLLDIMKSMR NLVKKYEPPR SEEVAVLKQK LERCHSAELA LNVITKKRKG YEDDDYVSKK SKHEEEEWT DDDLVESL UniProtKB: Cyclin-H |
-Macromolecule #3: Cyclin-dependent kinase 7
| Macromolecule | Name: Cyclin-dependent kinase 7 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: cyclin-dependent kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.362598 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SNAMALDVKS RAKRYEKLDF LGEGQFATVY KARDKNTNQI VAIKKIKLGH RSEAKDGINR TALREIKLLQ ELSHPNIIGL LDAFGHKSN ISLVFDFMET DLEVIIKDNS LVLTPSHIKA YMLMTLQGLE YLHQHWILHR DLKPNNLLLD ENGVLKLADF G LAKSFGSP ...String: SNAMALDVKS RAKRYEKLDF LGEGQFATVY KARDKNTNQI VAIKKIKLGH RSEAKDGINR TALREIKLLQ ELSHPNIIGL LDAFGHKSN ISLVFDFMET DLEVIIKDNS LVLTPSHIKA YMLMTLQGLE YLHQHWILHR DLKPNNLLLD ENGVLKLADF G LAKSFGSP NRAYTHQVVT RWYRAPELLF GARMYGVGVD MWAVGCILAE LLLRVPFLPG DSDLDQLTRI FETLGTPTEE QW PDMCSLP DYVTFKSFPG IPLHHIFSAA GDDLLDLIQG LFLFNPCARI TATQALKMKY FSNRPGPTPG CQLPRPNCPV ETL KEQSNP ALAIKRKRTE ALEQGGLPKK LIF UniProtKB: Cyclin-dependent kinase 7 |
-Macromolecule #4: 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,...
| Macromolecule | Name: 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium type: ligand / ID: 4 / Number of copies: 1 / Formula: 1QK |
|---|---|
| Molecular weight | Theoretical: 397.494 Da |
| Chemical component information | 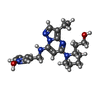 ChemComp-1QK: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 170 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 50 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number grids imaged: 1 / Number real images: 5210 / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 245614 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.4 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)