[English] 日本語
 Yorodumi
Yorodumi- EMDB-17382: Structure of human SIT1:ACE2 complex (closed PD conformation) bou... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate | |||||||||
 Map data Map data | cryoSPARC NU refinement map sharpened with a bfactor of -75 used for model refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | amino acid transporter / proline / SLC6A20 / ACE2 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationproline:sodium symporter activity / L-isoleucine transmembrane transporter activity / solute:sodium symporter activity / L-isoleucine import across plasma membrane / L-proline import across plasma membrane / Variant SLC6A20 contributes towards hyperglycinuria (HG) and iminoglycinuria (IG) / Variant SLC6A20 contributes towards hyperglycinuria (HG) and iminoglycinuria (IG) / amino-acid betaine transport / proline import across plasma membrane / L-proline transmembrane transporter activity ...proline:sodium symporter activity / L-isoleucine transmembrane transporter activity / solute:sodium symporter activity / L-isoleucine import across plasma membrane / L-proline import across plasma membrane / Variant SLC6A20 contributes towards hyperglycinuria (HG) and iminoglycinuria (IG) / Variant SLC6A20 contributes towards hyperglycinuria (HG) and iminoglycinuria (IG) / amino-acid betaine transport / proline import across plasma membrane / L-proline transmembrane transporter activity / amino-acid betaine transmembrane transporter activity / glycine import across plasma membrane / glycine transport / amino acid import across plasma membrane / proline transport / neutral L-amino acid transmembrane transporter activity / amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / Na+/Cl- dependent neurotransmitter transporters / amino acid transport / positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / angiotensin-mediated drinking behavior / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / regulation of systemic arterial blood pressure by renin-angiotensin / positive regulation of gap junction assembly / tryptophan transport / regulation of cardiac conduction / regulation of vasoconstriction / peptidyl-dipeptidase activity / maternal process involved in female pregnancy / transport across blood-brain barrier / Metabolism of Angiotensinogen to Angiotensins / carboxypeptidase activity / angiotensin maturation / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / metallocarboxypeptidase activity / viral life cycle / sodium ion transmembrane transport / positive regulation of cardiac muscle contraction / regulation of cytokine production / regulation of transmembrane transporter activity / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / metallopeptidase activity / positive regulation of reactive oxygen species metabolic process / endocytic vesicle membrane / regulation of cell population proliferation / virus receptor activity / regulation of inflammatory response / endopeptidase activity / Potential therapeutics for SARS / Induction of Cell-Cell Fusion / entry receptor-mediated virion attachment to host cell / membrane fusion / Attachment and Entry / receptor-mediated virion attachment to host cell / cilium / apical plasma membrane / membrane raft / endoplasmic reticulum lumen / symbiont entry into host cell / cell surface / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
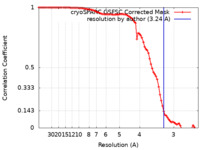
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Li HZ / Pike ACW / Chi G / Hansen JS / Lee SG / Rodstrom KEJ / Bushell SR / Speedman D / Evans A / Wang D ...Li HZ / Pike ACW / Chi G / Hansen JS / Lee SG / Rodstrom KEJ / Bushell SR / Speedman D / Evans A / Wang D / He D / Shrestha L / Nasrallah C / Chalk R / Moreira T / MacLean EM / Marsden B / Bountra C / Burgess-Brown NA / Dafforn TR / Carpenter EP / Sauer DB | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2. Authors: Huanyu Z Li / Ashley C W Pike / Irina Lotsaris / Gamma Chi / Jesper S Hansen / Sarah C Lee / Karin E J Rödström / Simon R Bushell / David Speedman / Adam Evans / Dong Wang / Didi He / ...Authors: Huanyu Z Li / Ashley C W Pike / Irina Lotsaris / Gamma Chi / Jesper S Hansen / Sarah C Lee / Karin E J Rödström / Simon R Bushell / David Speedman / Adam Evans / Dong Wang / Didi He / Leela Shrestha / Chady Nasrallah / Nicola A Burgess-Brown / Robert J Vandenberg / Timothy R Dafforn / Elisabeth P Carpenter / David B Sauer /   Abstract: Proline is widely known as the only proteogenic amino acid with a secondary amine. In addition to its crucial role in protein structure, the secondary amino acid modulates neurotransmission and ...Proline is widely known as the only proteogenic amino acid with a secondary amine. In addition to its crucial role in protein structure, the secondary amino acid modulates neurotransmission and regulates the kinetics of signaling proteins. To understand the structural basis of proline import, we solved the structure of the proline transporter SIT1 in complex with the COVID-19 viral receptor ACE2 by cryo-electron microscopy. The structure of pipecolate-bound SIT1 reveals the specific sequence requirements for proline transport in the SLC6 family and how this protein excludes amino acids with extended side chains. By comparing apo and substrate-bound SIT1 states, we also identify the structural changes that link substrate release and opening of the cytoplasmic gate and provide an explanation for how a missense mutation in the transporter causes iminoglycinuria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17382.map.gz emd_17382.map.gz | 33 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17382-v30.xml emd-17382-v30.xml emd-17382.xml emd-17382.xml | 26.1 KB 26.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17382_fsc.xml emd_17382_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17382.png emd_17382.png | 163.2 KB | ||
| Filedesc metadata |  emd-17382.cif.gz emd-17382.cif.gz | 8.2 KB | ||
| Others |  emd_17382_additional_1.map.gz emd_17382_additional_1.map.gz emd_17382_half_map_1.map.gz emd_17382_half_map_1.map.gz emd_17382_half_map_2.map.gz emd_17382_half_map_2.map.gz | 31.9 MB 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17382 http://ftp.pdbj.org/pub/emdb/structures/EMD-17382 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17382 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17382 | HTTPS FTP |
-Validation report
| Summary document |  emd_17382_validation.pdf.gz emd_17382_validation.pdf.gz | 861.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17382_full_validation.pdf.gz emd_17382_full_validation.pdf.gz | 860.7 KB | Display | |
| Data in XML |  emd_17382_validation.xml.gz emd_17382_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_17382_validation.cif.gz emd_17382_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17382 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17382 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17382 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17382 | HTTPS FTP |
-Related structure data
| Related structure data |  8p31MC  8p2wC  8p2xC  8p2yC  8p2zC  8p30C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17382.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17382.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC NU refinement map sharpened with a bfactor of -75 used for model refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2465 Å | ||||||||||||||||||||||||||||||||||||
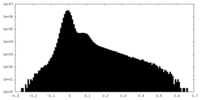
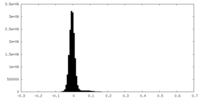
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened NU refinement full map
| File | emd_17382_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened NU refinement full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
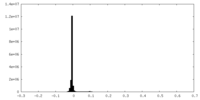
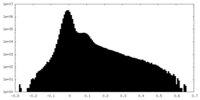
| Density Histograms |
-Half map: half map2
| File | emd_17382_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
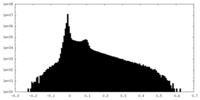
| Density Histograms |
-Half map: half map1
| File | emd_17382_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
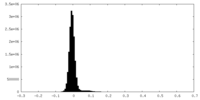
| Density Histograms |
- Sample components
Sample components
-Entire : SIT1:ACE2 complex
| Entire | Name: SIT1:ACE2 complex |
|---|---|
| Components |
|
-Supramolecule #1: SIT1:ACE2 complex
| Supramolecule | Name: SIT1:ACE2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 340 KDa |
-Macromolecule #1: Processed angiotensin-converting enzyme 2
| Macromolecule | Name: Processed angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.42557 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSSSSWLLLS LVAVTAADYK DDDDKSTIEE QAKTFLDKFN HEAEDLFYQS SLASWNYNTN ITEENVQNMN NAGDKWSAFL KEQSTLAQM YPLQEIQNLT VKLQLQALQQ NGSSVLSEDK SKRLNTILNT MSTIYSTGKV CNPDNPQECL LLEPGLNEIM A NSLDYNER ...String: MSSSSWLLLS LVAVTAADYK DDDDKSTIEE QAKTFLDKFN HEAEDLFYQS SLASWNYNTN ITEENVQNMN NAGDKWSAFL KEQSTLAQM YPLQEIQNLT VKLQLQALQQ NGSSVLSEDK SKRLNTILNT MSTIYSTGKV CNPDNPQECL LLEPGLNEIM A NSLDYNER LWAWESWRSE VGKQLRPLYE EYVVLKNEMA RANHYEDYGD YWRGDYEVNG VDGYDYSRGQ LIEDVEHTFE EI KPLYEHL HAYVRAKLMN AYPSYISPIG CLPAHLLGDM WGRFWTNLYS LTVPFGQKPN IDVTDAMVDQ AWDAQRIFKE AEK FFVSVG LPNMTQGFWE NSMLTDPGNV QKAVCHPTAW DLGKGDFRIL MCTKVTMDDF LTAHHEMGHI QYDMAYAAQP FLLR NGANE GFHEAVGEIM SLSAATPKHL KSIGLLSPDF QEDNETEINF LLKQALTIVG TLPFTYMLEK WRWMVFKGEI PKDQW MKKW WEMKREIVGV VEPVPHDETY CDPASLFHVS NDYSFIRYYT RTLYQFQFQE ALCQAAKHEG PLHKCDISNS TEAGQK LFN MLRLGKSEPW TLALENVVGA KNMNVRPLLN YFEPLFTWLK DQNKNSFVGW STDWSPYADQ SIKVRISLKS ALGDKAY EW NDNEMYLFRS SVAYAMRQYF LKVKNQMILF GEEDVRVANL KPRISFNFFV TAPKNVSDII PRTEVEKAIR MSRSRIND A FRLNDNSLEF LGIQPTLGPP NQPPVSIWLI VFGVVMGVIV VGIVILIFTG IRDRKKKNKA RSGENPYASI DISKGENNP GFQNTDDVQT SF UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #2: Sodium- and chloride-dependent transporter XTRP3
| Macromolecule | Name: Sodium- and chloride-dependent transporter XTRP3 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.376258 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEKARPLWAN SLQFVFACIS YAVGLGNVWR FPYLCQMYGG GSFLVPYIIM LIVEGMPLLY LELAVGQRMR QGSIGAWRTI SPYLSGVGV ASVVVSFFLS MYYNVINAWA FWYLFHSFQD PLPWSVCPLN GNHTGYDEEC EKASSTQYFW YRKTLNISPS L QENGGVQW ...String: MEKARPLWAN SLQFVFACIS YAVGLGNVWR FPYLCQMYGG GSFLVPYIIM LIVEGMPLLY LELAVGQRMR QGSIGAWRTI SPYLSGVGV ASVVVSFFLS MYYNVINAWA FWYLFHSFQD PLPWSVCPLN GNHTGYDEEC EKASSTQYFW YRKTLNISPS L QENGGVQW EPALCLLLAW LVVYLCILRG TESTGKVVYF TASLPYCVLI IYLIRGLTLH GATNGLMYMF TPKIEQLANP KA WINAATQ IFFSLGLGFG SLIAFASYNE PSNNCQKHAI IVSLINSFTS IFASIVTFSI YGFKATFNYE NCLKKVSLLL TNT FDLEDG FLTASNLEQV KGYLASAYPS KYSEMFPQIK NCSLESELDT AVQGTGLAFI VYTEAIKNME VSQLWSVLYF FMLL MLGIG SMLGNTAAIL TPLTDSKIIS SHLPKEAISG LVCLVNCAIG MVFTMEAGNY WFDIFNDYAA TLSLLLIVLV ETIAV CYVY GLRRFESDLK AMTGRAVSWY WKVMWAGVSP LLIVSLFVFY LSDYILTGTL KYQAWDASQG QLVTKDYPAY ALAVIG LLV ASSTMCIPLA ALGTFVQRRL KRGDADPVAA ENLYFQSHHH HHHHHHHGSA WSHPQFEKGG GSGGGSGGSA WSHPQFE K UniProtKB: Sodium- and chloride-dependent transporter XTRP3 |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: 2-acetamido-2-deoxy-alpha-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-alpha-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 2 / Formula: NDG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NDG: |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: (2S)-piperidine-2-carboxylic acid
| Macromolecule | Name: (2S)-piperidine-2-carboxylic acid / type: ligand / ID: 8 / Number of copies: 2 / Formula: YCP |
|---|---|
| Molecular weight | Theoretical: 129.157 Da |
| Chemical component information | 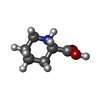 ChemComp-YCP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: grid blotted for 4sec after a 20sec wait time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Software | Name: EPU |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 13194 / Average exposure time: 1.77 sec. / Average electron dose: 50.0 e/Å2 Details: Images were collected in super-resolution mode EPU bin2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: C / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Software | Name:  Coot (ver. 0.9.6) Coot (ver. 0.9.6) |
| Details | Initial model fitting was peformed in Chimera and model building in COOT |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 75 |
| Output model |  PDB-8p31: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)