[English] 日本語
 Yorodumi
Yorodumi- EMDB-17266: ATM(Q2971A) dimeric C-terminal region activated by oxidative stre... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | ATM(Q2971A) dimeric C-terminal region activated by oxidative stress in complex with Mg AMP-PNP and p53 peptide | ||||||||||||
 Map data Map data | Unsharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Kinase / Ataxia-Telangiectasia Mutated / ATM / p53 / SIGNALING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of DNA catabolic process / establishment of RNA localization to telomere / positive regulation of telomerase catalytic core complex assembly / positive regulation of DNA damage response, signal transduction by p53 class mediator / cellular response to nitrosative stress / establishment of protein-containing complex localization to telomere / negative regulation of telomere capping / Sensing of DNA Double Strand Breaks / positive regulation of telomere maintenance via telomere lengthening / regulation of microglial cell activation ...positive regulation of DNA catabolic process / establishment of RNA localization to telomere / positive regulation of telomerase catalytic core complex assembly / positive regulation of DNA damage response, signal transduction by p53 class mediator / cellular response to nitrosative stress / establishment of protein-containing complex localization to telomere / negative regulation of telomere capping / Sensing of DNA Double Strand Breaks / positive regulation of telomere maintenance via telomere lengthening / regulation of microglial cell activation / meiotic telomere clustering / pre-B cell allelic exclusion / DNA-dependent protein kinase activity / histone H2AXS139 kinase activity / male meiotic nuclear division / histone mRNA catabolic process / female meiotic nuclear division / pexophagy / cellular response to X-ray / regulation of telomere maintenance via telomerase / peptidyl-serine autophosphorylation / DNA double-strand break processing / lipoprotein catabolic process / V(D)J recombination / regulation of autophagosome assembly / oocyte development / Impaired BRCA2 binding to PALB2 / reciprocal meiotic recombination / Loss of function of TP53 in cancer due to loss of tetramerization ability / Regulation of TP53 Expression / signal transduction by p53 class mediator / negative regulation of G1 to G0 transition / negative regulation of glucose catabolic process to lactate via pyruvate / Transcriptional activation of cell cycle inhibitor p21 / regulation of intrinsic apoptotic signaling pathway by p53 class mediator / Activation of NOXA and translocation to mitochondria / negative regulation of pentose-phosphate shunt / ATP-dependent DNA/DNA annealing activity / negative regulation of helicase activity / regulation of cell cycle G2/M phase transition / intrinsic apoptotic signaling pathway in response to hypoxia / regulation of fibroblast apoptotic process / oxidative stress-induced premature senescence / oligodendrocyte apoptotic process / negative regulation of miRNA processing / positive regulation of thymocyte apoptotic process / glucose catabolic process to lactate via pyruvate / regulation of tissue remodeling / DNA repair complex / positive regulation of mitochondrial membrane permeability / negative regulation of mitophagy / positive regulation of programmed necrotic cell death / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Homologous DNA Pairing and Strand Exchange / mRNA transcription / bone marrow development / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / circadian behavior / histone deacetylase regulator activity / germ cell nucleus / regulation of mitochondrial membrane permeability involved in apoptotic process / RUNX3 regulates CDKN1A transcription / regulation of DNA damage response, signal transduction by p53 class mediator / Resolution of D-loop Structures through Holliday Junction Intermediates / TP53 regulates transcription of additional cell cycle genes whose exact role in the p53 pathway remain uncertain / TP53 Regulates Transcription of Death Receptors and Ligands / Activation of PUMA and translocation to mitochondria / DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator / HDR through Single Strand Annealing (SSA) / negative regulation of glial cell proliferation / negative regulation of neuroblast proliferation / Impaired BRCA2 binding to RAD51 / Regulation of TP53 Activity through Association with Co-factors / 1-phosphatidylinositol-3-kinase activity / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / mitochondrial DNA repair / T cell lineage commitment / TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest / response to ionizing radiation / ER overload response / mitotic spindle assembly checkpoint signaling / negative regulation of DNA replication / B cell lineage commitment / positive regulation of cardiac muscle cell apoptotic process / thymocyte apoptotic process / TP53 regulates transcription of several additional cell death genes whose specific roles in p53-dependent apoptosis remain uncertain / TP53 Regulates Transcription of Caspase Activators and Caspases / mitotic G1 DNA damage checkpoint signaling / cardiac septum morphogenesis / negative regulation of B cell proliferation / positive regulation of execution phase of apoptosis / entrainment of circadian clock by photoperiod / PI5P Regulates TP53 Acetylation / Association of TriC/CCT with target proteins during biosynthesis / mitotic G2 DNA damage checkpoint signaling / Zygotic genome activation (ZGA) / necroptotic process / positive regulation of release of cytochrome c from mitochondria Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Howes AC / Perisic O / Williams RL | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural insights into the activation of ataxia-telangiectasia mutated by oxidative stress. Authors: Anna C Howes / Olga Perisic / Roger L Williams /  Abstract: Ataxia-telangiectasia mutated (ATM) is a master kinase regulating DNA damage response that is activated by DNA double-strand breaks. However, ATM is also directly activated by reactive oxygen ...Ataxia-telangiectasia mutated (ATM) is a master kinase regulating DNA damage response that is activated by DNA double-strand breaks. However, ATM is also directly activated by reactive oxygen species, but how oxidative activation is achieved remains unknown. We determined the cryo-EM structure of an HO-activated ATM and showed that under oxidizing conditions, ATM formed an intramolecular disulfide bridge between two protomers that are rotated relative to each other when compared to the basal state. This rotation is accompanied by release of the substrate-blocking PRD region and twisting of the N-lobe relative to the C-lobe, which greatly optimizes catalysis. This active site remodeling enabled us to capture a substrate (p53) bound to the enzyme. This provides the first structural insights into how ATM is activated during oxidative stress. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17266.map.gz emd_17266.map.gz | 121.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17266-v30.xml emd-17266-v30.xml emd-17266.xml emd-17266.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17266_fsc.xml emd_17266_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17266.png emd_17266.png | 85.2 KB | ||
| Masks |  emd_17266_msk_1.map emd_17266_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17266.cif.gz emd-17266.cif.gz | 8.2 KB | ||
| Others |  emd_17266_additional_1.map.gz emd_17266_additional_1.map.gz emd_17266_half_map_1.map.gz emd_17266_half_map_1.map.gz emd_17266_half_map_2.map.gz emd_17266_half_map_2.map.gz | 230 MB 226.1 MB 226.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17266 http://ftp.pdbj.org/pub/emdb/structures/EMD-17266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17266 | HTTPS FTP |
-Related structure data
| Related structure data |  8oxoMC  8oxmC  8oxpC  8oxqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17266.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17266.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17266_msk_1.map emd_17266_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
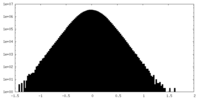
| Density Histograms |
-Additional map: Sharpened map
| File | emd_17266_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
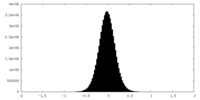
| Density Histograms |
-Half map: Half Map A
| File | emd_17266_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
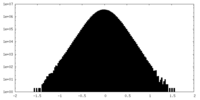
| Density Histograms |
-Half map: Half Map B
| File | emd_17266_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATM(Q2971A) dimeric C-terminal region activated by H2O2 and bound...
| Entire | Name: ATM(Q2971A) dimeric C-terminal region activated by H2O2 and bound to Mg AMP-PNP and p53 substrate peptide |
|---|---|
| Components |
|
-Supramolecule #1: ATM(Q2971A) dimeric C-terminal region activated by H2O2 and bound...
| Supramolecule | Name: ATM(Q2971A) dimeric C-terminal region activated by H2O2 and bound to Mg AMP-PNP and p53 substrate peptide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Only the C-terminal region of the ATM (Q2971A) dimer from local refinement is shown, with AMP-PNP and p53 substrate peptide bound in the structure. Please note, the whole dimer molecule is present in the sample. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cellular tumor antigen p53
| Macromolecule | Name: Cellular tumor antigen p53 / type: protein_or_peptide / ID: 1 / Details: Custom synthesized peptide / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.362438 KDa |
| Sequence | String: EPPLSQETFS DL UniProtKB: Cellular tumor antigen p53 |
-Macromolecule #2: Serine-protein kinase ATM
| Macromolecule | Name: Serine-protein kinase ATM / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 365.005562 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKH MGVQVETISP GDGRTFPKRG QTCVVHYTGM LEDGKKFDSS RDRNKPFKFM LGKQEVIRGW EEGVAQMSVG QRAKLTISP DYAYGATGHP GIIPPHATLV FDVELLKLEG GSAGSGSASM SLVLNDLLIC CRQLEHDRAT ERKKEVEKFK R LIRDPETI ...String: MDYKDDDDKH MGVQVETISP GDGRTFPKRG QTCVVHYTGM LEDGKKFDSS RDRNKPFKFM LGKQEVIRGW EEGVAQMSVG QRAKLTISP DYAYGATGHP GIIPPHATLV FDVELLKLEG GSAGSGSASM SLVLNDLLIC CRQLEHDRAT ERKKEVEKFK R LIRDPETI KHLDRHSDSK QGKYLNWDAV FRFLQKYIQK ETECLRIAKP NVSASTQASR QKKMQEISSL VKYFIKCANR RA PRLKCQE LLNYIMDTVK DSSNGAIYGA DCSNILLKDI LSVRKYWCEI SQQQWLELFS VYFRLYLKPS QDVHRVLVAR IIH AVTKGC CSQTDGLNSK FLDFFSKAIQ CARQEKSSSG LNHILAALTI FLKTLAVNFR IRVCELGDEI LPTLLYIWTQ HRLN DSLKE VIIELFQLQI YIHHPKGAKT QEKGAYESTK WRSILYNLYD LLVNEISHIG SRGKYSSGFR NIAVKENLIE LMADI CHQV FNEDTRSLEI SQSYTTTQRE SSDYSVPCKR KKIELGWEVI KDHLQKSQND FDLVPWLQIA TQLISKYPAS LPNCEL SPL LMILSQLLPQ QRHGERTPYV LRCLTEVALC QDKRSNLESS QKSDLLKLWN KIWCITFRGI SSEQIQAENF GLLGAII QG SLVEVDREFW KLFTGSACRP SCPAVCCLTL ALTTSIVPGT VKMGIEQNMC EVNRSFSLKE SIMKWLLFYQ LEGDLENS T EVPPILHSNF PHLVLEKILV SLTMKNCKAA MNFFQSVPEC EHHQKDKEEL SFSEVEELFL QTTFDKMDFL TIVRECGIE KHQSSIGFSV HQNLKESLDR CLLGLSEQLL NNYSSEITNS ETLVRCSRLL VGVLGCYCYM GVIAEEEAYK SELFQKAKSL MQCAGESIT LFKNKTNEEF RIGSLRNMMQ LCTRCLSNCT KKSPNKIASG FFLRLLTSKL MNDIADICKS LASFIKKPFD R GEVESMED DTNGNLMEVE DQSSMNLFND YPDSSVSDAN EPGESQSTIG AINPLAEEYL SKQDLLFLDM LKFLCLCVTT AQ TNTVSFR AADIRRKLLM LIDSSTLEPT KSLHLHMYLM LLKELPGEEY PLPMEDVLEL LKPLSNVCSL YRRDQDVCKT ILN HVLHVV KNLGQSNMDS ENTRDAQGQF LTVIGAFWHL TKERKYIFSV RMALVNCLKT LLEADPYSKW AILNVMGKDF PVNE VFTQF LADNHHQVRM LAAESINRLF QDTKGDSSRL LKALPLKLQQ TAFENAYLKA QEGMREMSHS AENPETLDEI YNRKS VLLT LIAVVLSCSP ICEKQALFAL CKSVKENGLE PHLVKKVLEK VSETFGYRRL EDFMASHLDY LVLEWLNLQD TEYNLS SFP FILLNYTNIE DFYRSCYKVL IPHLVIRSHF DEVKSIANQI QEDWKSLLTD CFPKILVNIL PYFAYEGTRD SGMAQQR ET ATKVYDMLKS ENLLGKQIDH LFISNLPEIV VELLMTLHEP ANSSASQSTD LCDFSGDLDP APNPPHFPSH VIKATFAY I SNCHKTKLKS ILEILSKSPD SYQKILLAIC EQAAETNNVY KKHRILKIYH LFVSLLLKDI KSGLGGAWAF VLRDVIYTL IHYINQRPSC IMDVSLRSFS LCCDLLSQVC QTAVTYCKDA LENHLHVIVG TLIPLVYEQV EVQKQVLDLL KYLVIDNKDN ENLYITIKL LDPFPDHVVF KDLRITQQKI KYSRGPFSLL EEINHFLSVS VYDALPLTRL EGLKDLRRQL ELHKDQMVDI M RASQDNPQ DGIMVKLVVN LLQLSKMAIN HTGEKEVLEA VGSCLGEVGP IDFSTIAIQH SKDASYTKAL KLFEDKELQW TF IMLTYLN NTLVEDCVKV RSAAVTCLKN ILATKTGHSF WEIYKMTTDP MLAYLQPFRT SRKKFLEVPR FDKENPFEGL DDI NLWIPL SENHDIWIKT LTCAFLDSGG TKCEILQLLK PMCEVKTDFC QTVLPYLIHD ILLQDTNESW RNLLSTHVQG FFTS CLRHF SQTSRSTTPA NLDSESEHFF RCCLDKKSQR TMLAVVDYMR RQKRPSSGTI FNDAFWLDLN YLEVAKVAQS CAAHF TALL YAEIYADKKS MDDQEKRSLA FEEGSQSTTI SSLSEKSKEE TGISLQDLLL EIYRSIGEPD SLYGCGGGKM LQPITR LRT YEHEAMWGKA LVTYDLETAI PSSTRQAGII QALQNLGLCH ILSVYLKGLD YENKDWCPEL EELHYQAAWR NMQWDHC TS VSKEVEGTSY HESLYNALQS LRDREFSTFY ESLKYARVKE VEEMCKRSLE SVYSLYPTLS RLQAIGELES IGELFSRS V THRQLSEVYI KWQKHSQLLK DSDFSFQEPI MALRTVILEI LMEKEMDNSQ RECIKDILTK HLVELSILAR TFKNTQLPE RAIFQIKQYN SVSCGVSEWQ LEEAQVFWAK KEQSLALSIL KQMIKKLDAS CAANNPSLKL TYTECLRVCG NWLAETCLEN PAVIMQTYL EKAVEVAGNY DGESSDELRN GKMKAFLSLA RFSDTQYQRI ENYMKSSEFE NKQALLKRAK EEVGLLREHK I QTNRYTVK VQRELELDEL ALRALKEDRK RFLCKAVENY INCLLSGEEH DMWVFRLCSL WLENSGVSEV NGMMKRDGMK IP TYKFLPL MYQLAARMGT KMMGGLGFHE VLNNLISRIS MDHPHHTLFI ILALANANRD EFLTKPEVAR RSRITKNVPK QSS QLDEDR TEAANRIICT IRSRRPQMVR SVEALCDAYI ILANLDATQW KTQRKGINIP ADQPITKLKN LEDVVVPTME IKVD HTGEY GNLVTIQSFK AEFRLAGGVN LPKIIDCVGS DGKERRQLVK GRDDLRQDAV MQQVFQMCNT LLQRNTETRK RKLTI CTYK VVPLSQRSGV LEWCTGTVPI GEFLVNNEDG AHKRYRPNDF SAFQCQKKMM EVQKKSFEEK YEVFMDVCQN FQPVFR YFC MEKFLDPAIW FEKRLAYTRS VATSSIVGYI LGLGDRHVQN ILINEQSAEL VHIDLGVAFE QGKILPTPET VPFRLTR DI VDGMGITGVE GVFRRCCEKT MEVMRNSQET LLTIVEVLLY DPLFDWTMNP LKALYLAQRP EDETELHPTL NADDQECK R NLSDIDQSFN KVAERVLMRL QEKLKGVEEG TVLSVGGQVN LLIQQAIDPK NLSRLFPGWK AWV UniProtKB: Serine-protein kinase ATM |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 39.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z
Z Y
Y X
X