+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DNA-unbound MutSbeta-ATP complex (bent clamp form) | |||||||||
 Map data Map data | DNA-unbound MutSbeta-ATP complex (bent clamp form) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA repair protein complex / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsomatic recombination of immunoglobulin genes involved in immune response / MutSbeta complex / Defective Mismatch Repair Associated With MSH3 / MutSalpha complex / Defective Mismatch Repair Associated With MSH2 / Defective Mismatch Repair Associated With MSH6 / guanine/thymine mispair binding / somatic recombination of immunoglobulin gene segments / B cell mediated immunity / maintenance of DNA repeat elements ...somatic recombination of immunoglobulin genes involved in immune response / MutSbeta complex / Defective Mismatch Repair Associated With MSH3 / MutSalpha complex / Defective Mismatch Repair Associated With MSH2 / Defective Mismatch Repair Associated With MSH6 / guanine/thymine mispair binding / somatic recombination of immunoglobulin gene segments / B cell mediated immunity / maintenance of DNA repeat elements / positive regulation of helicase activity / positive regulation of isotype switching to IgA isotypes / centromeric DNA binding / mitotic recombination / positive regulation of isotype switching to IgG isotypes / mismatched DNA binding / negative regulation of DNA recombination / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / isotype switching / oxidative phosphorylation / response to UV-B / postreplication repair / mitotic intra-S DNA damage checkpoint signaling / ATP-dependent DNA damage sensor activity / germ cell development / response to X-ray / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / somatic hypermutation of immunoglobulin genes / mismatch repair / ATP-dependent activity, acting on DNA / protein localization to chromatin / B cell differentiation / determination of adult lifespan / TP53 Regulates Transcription of DNA Repair Genes / male gonad development / double-strand break repair / double-stranded DNA binding / in utero embryonic development / negative regulation of neuron apoptotic process / damaged DNA binding / chromosome, telomeric region / DNA repair / chromatin binding / enzyme binding / protein homodimerization activity / ATP hydrolysis activity / DNA binding / nucleoplasm / ATP binding / membrane / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.43 Å | |||||||||
 Authors Authors | Lee J-H / Thomsen M / Daub H / Steinbacher S / Sztyler A / Thieulin-Pardo G / Neudegger T / Plotnikov N / Iyer RR / Wilkinson H ...Lee J-H / Thomsen M / Daub H / Steinbacher S / Sztyler A / Thieulin-Pardo G / Neudegger T / Plotnikov N / Iyer RR / Wilkinson H / Monteagudo E / Felsenfeld DP / Haque T / Finley M / Dominguez C / Vogt TF / Prasad BC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: DNA-unbound MutSbeta-ATP complex (bent clamp form) Authors: Lee J-H / Thomsen M / Daub H / Steinbacher S / Sztyler A / Thieulin-Pardo G / Neudegger T / Plotnikov N / Iyer RR / Wilkinson H / Monteagudo E / Felsenfeld DP / Haque T / Finley M / ...Authors: Lee J-H / Thomsen M / Daub H / Steinbacher S / Sztyler A / Thieulin-Pardo G / Neudegger T / Plotnikov N / Iyer RR / Wilkinson H / Monteagudo E / Felsenfeld DP / Haque T / Finley M / Dominguez C / Vogt TF / Prasad BC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16973.map.gz emd_16973.map.gz | 111.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16973-v30.xml emd-16973-v30.xml emd-16973.xml emd-16973.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16973.png emd_16973.png | 20 KB | ||
| Filedesc metadata |  emd-16973.cif.gz emd-16973.cif.gz | 6.7 KB | ||
| Others |  emd_16973_half_map_1.map.gz emd_16973_half_map_1.map.gz emd_16973_half_map_2.map.gz emd_16973_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16973 http://ftp.pdbj.org/pub/emdb/structures/EMD-16973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16973 | HTTPS FTP |
-Validation report
| Summary document |  emd_16973_validation.pdf.gz emd_16973_validation.pdf.gz | 590.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16973_full_validation.pdf.gz emd_16973_full_validation.pdf.gz | 590.3 KB | Display | |
| Data in XML |  emd_16973_validation.xml.gz emd_16973_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_16973_validation.cif.gz emd_16973_validation.cif.gz | 16.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16973 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16973 | HTTPS FTP |
-Related structure data
| Related structure data |  8omoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16973.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16973.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DNA-unbound MutSbeta-ATP complex (bent clamp form) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9142 Å | ||||||||||||||||||||||||||||||||||||
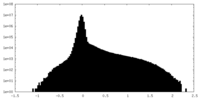
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: DNA-unbound MutSbeta-ATP complex (bent clamp form)
| File | emd_16973_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DNA-unbound MutSbeta-ATP complex (bent clamp form) | ||||||||||||
| Projections & Slices |
| ||||||||||||
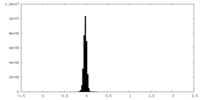
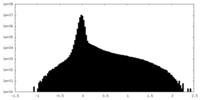
| Density Histograms |
-Half map: DNA-unbound MutSbeta-ATP complex (bent clamp form)
| File | emd_16973_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DNA-unbound MutSbeta-ATP complex (bent clamp form) | ||||||||||||
| Projections & Slices |
| ||||||||||||
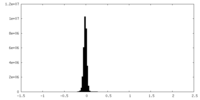
| Density Histograms |
- Sample components
Sample components
-Entire : DNA-unbound MutSbeta-ATP complex (bent clamp form)
| Entire | Name: DNA-unbound MutSbeta-ATP complex (bent clamp form) |
|---|---|
| Components |
|
-Supramolecule #1: DNA-unbound MutSbeta-ATP complex (bent clamp form)
| Supramolecule | Name: DNA-unbound MutSbeta-ATP complex (bent clamp form) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: DNA mismatch repair protein Msh2
| Macromolecule | Name: DNA mismatch repair protein Msh2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 104.861875 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAVQPKETLQ LESAAEVGFV RFFQGMPEKP TTTVRLFDRG DFYTAHGEDA LLAAREVFKT QGVIKYMGPA GAKNLQSVVL SKMNFESFV KDLLLVRQYR VEVYKNRAGN KASKENDWYL AYKASPGNLS QFEDILFGNN DMSASIGVVG VKMSAVDGQR Q VGVGYVDS ...String: MAVQPKETLQ LESAAEVGFV RFFQGMPEKP TTTVRLFDRG DFYTAHGEDA LLAAREVFKT QGVIKYMGPA GAKNLQSVVL SKMNFESFV KDLLLVRQYR VEVYKNRAGN KASKENDWYL AYKASPGNLS QFEDILFGNN DMSASIGVVG VKMSAVDGQR Q VGVGYVDS IQRKLGLCEF PDNDQFSNLE ALLIQIGPKE CVLPGGETAG DMGKLRQIIQ RGGILITERK KADFSTKDIY QD LNRLLKG KKGEQMNSAV LPEMENQVAV SSLSAVIKFL ELLSDDSNFG QFELTTFDFS QYMKLDIAAV RALNLFQGSV EDT TGSQSL AALLNKCKTP QGQRLVNQWI KQPLMDKNRI EERLNLVEAF VEDAELRQTL QEDLLRRFPD LNRLAKKFQR QAAN LQDCY RLYQGINQLP NVIQALEKHE GKHQKLLLAV FVTPLTDLRS DFSKFQEMIE TTLDMDQVEN HEFLVKPSFD PNLSE LREI MNDLEKKMQS TLISAARDLG LDPGKQIKLD SSAQFGYYFR VTCKEEKVLR NNKNFSTVDI QKNGVKFTNS KLTSLN EEY TKNKTEYEEA QDAIVKEIVN ISSGYVEPMQ TLNDVLAQLD AVVSFAHVSN GAPVPYVRPA ILEKGQGRII LKASRHA CV EVQDEIAFIP NDVYFEKDKQ MFHIITGPNM GGKSTYIRQT GVIVLMAQIG CFVPCESAEV SIVDCILARV GAGDSQLK G VSTFMAEMLE TASILRSATK DSLIIIDELG RGTSTYDGFG LAWAISEYIA TKIGAFCMFA THFHELTALA NQIPTVNNL HVTALTTEET LTMLYQVKKG VCDQSFGIHV AELANFPKHV IECAKQKALE LEEFQYIGES QGYDIMEPAA KKCYLEREQG EKIIQEFLS KVKQMPFTEM SEENITIKLK QLKAEVIAKN NSFVNEIISR IKVTT UniProtKB: DNA mismatch repair protein Msh2 |
-Macromolecule #2: DNA mismatch repair protein Msh3
| Macromolecule | Name: DNA mismatch repair protein Msh3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.618703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRRKPASGG LAASSSAPAR QAVLSRFFQS TGSLKSTSSS TGAADQVDPG AAAAAAAAAA AAPPAPPAPA FPPQLPPHIA TEIDRRKKR PLENDGPVKK KVKKVQQKEG GSDLGMSGNS EPKKCLRTRN VSKSLEKLKE FCCDSALPQS RVQTESLQER F AVLPKCTD ...String: MSRRKPASGG LAASSSAPAR QAVLSRFFQS TGSLKSTSSS TGAADQVDPG AAAAAAAAAA AAPPAPPAPA FPPQLPPHIA TEIDRRKKR PLENDGPVKK KVKKVQQKEG GSDLGMSGNS EPKKCLRTRN VSKSLEKLKE FCCDSALPQS RVQTESLQER F AVLPKCTD FDDISLLHAK NAVSSEDSKR QINQKDTTLF DLSQFGSSNT SHENLQKTAS KSANKRSKSI YTPLELQYIE MK QQHKDAV LCVECGYKYR FFGEDAEIAA RELNIYCHLD HNFMTASIPT HRLFVHVRRL VAKGYKVGVV KQTETAALKA IGD NRSSLF SRKLTALYTK STLIGEDVNP LIKLDDAVNV DEIMTDTSTS YLLCISENKE NVRDKKKGNI FIGIVGVQPA TGEV VFDSF QDSASRSELE TRMSSLQPVE LLLPSALSEQ TEALIHRATS VSVQDDRIRV ERMDNIYFEY SHAFQAVTEF YAKDT VDIK GSQIISGIVN LEKPVICSLA AIIKYLKEFN LEKMLSKPEN FKQLSSKMEF MTINGTTLRN LEILQNQTDM KTKGSL LWV LDHTKTSFGR RKLKKWVTQP LLKLREINAR LDAVSEVLHS ESSVFGQIEN HLRKLPDIER GLCSIYHKKC STQEFFL IV KTLYHLKSEF QAIIPAVNSH IQSDLLRTVI LEIPELLSPV EHYLKILNEQ AAKVGDKTEL FKDLSDFPLI KKRKDEIQ G VIDEIRMHLQ EIRKILKNPS AQYVTVSGQE FMIEIKNSAV SCIPTDWVKV GSTKAVSRFH SPFIVENYRH LNQLREQLV LDCSAEWLDF LEKFSEHYHS LCKAVHHLAT VDCIFSLAKV AKQGDYCRPT VQEERKIVIK NGRHPVIDVL LGEQDQYVPN NTDLSEDSE RVMIITGPNM GGKSSYIKQV ALITIMAQIG SYVPAEEATI GIVDGIFTRM GAADNIYKGR STFMEELTDT A EIIRKATS QSLVILDELG RGTSTHDGIA IAYATLEYFI RDVKSLTLFV THYPPVCELE KNYSHQVGNY HMGFLVSEDE SK LDPGAAE QVPDFVTFLY QITRGIAARS YGLNVAKLAD VPGEILKKAA HKSKELEGLI NTKRKRLKYF AKLWTMHNAQ DLQ KWTEEF NMEETQTSLL H UniProtKB: DNA mismatch repair protein Msh3 |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 57.38 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.43 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 263343 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)