+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM KSB domain of RhiE from Burkholderia rhizoxinica | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Polyketide / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationDIM/DIP cell wall layer assembly / fatty acid synthase activity / secondary metabolite biosynthetic process / phosphopantetheine binding / 3-oxoacyl-[acyl-carrier-protein] synthase activity / fatty acid biosynthetic process / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Mycetohabitans rhizoxinica (bacteria) Mycetohabitans rhizoxinica (bacteria) | |||||||||
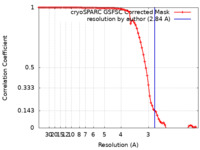
| Method | single particle reconstruction / cryo EM / Resolution: 2.84 Å | |||||||||
 Authors Authors | Capper MJ / Koehnke J | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural Analysis of a Chain-Branching Polyketide Synthase Module Authors: Dell M / Tran MA / Capper MJ / Sundaram S / Fiedler J / Koehnke J / Hellmich U / Hertwick C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16892.map.gz emd_16892.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16892-v30.xml emd-16892-v30.xml emd-16892.xml emd-16892.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16892_fsc.xml emd_16892_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_16892.png emd_16892.png | 149.1 KB | ||
| Masks |  emd_16892_msk_1.map emd_16892_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16892.cif.gz emd-16892.cif.gz | 6.5 KB | ||
| Others |  emd_16892_additional_1.map.gz emd_16892_additional_1.map.gz emd_16892_half_map_1.map.gz emd_16892_half_map_1.map.gz emd_16892_half_map_2.map.gz emd_16892_half_map_2.map.gz | 56.2 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16892 http://ftp.pdbj.org/pub/emdb/structures/EMD-16892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16892 | HTTPS FTP |
-Validation report
| Summary document |  emd_16892_validation.pdf.gz emd_16892_validation.pdf.gz | 901.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16892_full_validation.pdf.gz emd_16892_full_validation.pdf.gz | 901 KB | Display | |
| Data in XML |  emd_16892_validation.xml.gz emd_16892_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_16892_validation.cif.gz emd_16892_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16892 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16892 | HTTPS FTP |
-Related structure data
| Related structure data |  8oiiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16892.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16892.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.015 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16892_msk_1.map emd_16892_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_16892_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16892_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16892_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimeric complex of RhiE KSB domains
| Entire | Name: Homodimeric complex of RhiE KSB domains |
|---|---|
| Components |
|
-Supramolecule #1: Homodimeric complex of RhiE KSB domains
| Supramolecule | Name: Homodimeric complex of RhiE KSB domains / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycetohabitans rhizoxinica (bacteria) Mycetohabitans rhizoxinica (bacteria) |
| Molecular weight | Theoretical: 230 KDa |
-Macromolecule #1: RhiE protein,Polyketide synthase domain protein RhiE
| Macromolecule | Name: RhiE protein,Polyketide synthase domain protein RhiE / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycetohabitans rhizoxinica (bacteria) Mycetohabitans rhizoxinica (bacteria) |
| Molecular weight | Theoretical: 115.527039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKHHHHHHHH GGLVPRGSHG GSSGERVEDN ELANYIAVIG LGGYYPGADS IDELWQNLAN GVDCMSDFPA DRWDHSKIYY KNRKVLGKT TCINGSFIKD VDKFDYSYFK MPKVYADHMS PEVRLFLQVA VHTFEDAGYS KETLLSRYNG DVGVLLGTMS N DYHYYGFE ...String: MKHHHHHHHH GGLVPRGSHG GSSGERVEDN ELANYIAVIG LGGYYPGADS IDELWQNLAN GVDCMSDFPA DRWDHSKIYY KNRKVLGKT TCINGSFIKD VDKFDYSYFK MPKVYADHMS PEVRLFLQVA VHTFEDAGYS KETLLSRYNG DVGVLLGTMS N DYHYYGFE SNVFRGSMAS GSGMATIPMT VSYFYGLTGP SLFIDTMCSS SSTCIHTACQ MLKHDETKMV LAGGLNLMYH PY TTVNTSQ GNFTSITSES VNSYGVGADG TVIGEGIGAV LLKRLDRAIA DRDQIYGVIK GSAMTNAGER NGFNVPNPDL QTL AIRQAM DQAKVHPSSI SYIEGHGSGT KLGDPIEVLG LNNAFRWATD DKQFCYLGSI KSNIGHLLAA SGIAGLTKTL LQFK HKQIA PSIHSSQLNQ DIDFADTPFV VPQQLIEWRQ PERIINGRKQ VFPRRAGLTS IAAGGMNAHM IVEEYPEPAD SAGQI SEDQ LVFVFSVHKL ALLAQNLTSF RDWLASSEAP LAQIAYTLQV GKNNLRNRLA IRCRTRQALS RALNACIDGH YQSSAD SKI FYRFQESDAV QPLESDLNDP LAPLLTQWLN GDSQVDWASL YAQPPVRISL PAYRFEKTRC WYTEEGYESS IVNPLMF KN KLHPLVAKNC STPQPGAIFR TDFVEDELLD YVYSGRGGRR LSAFNFADVA LAMPALASRF DGRTLSVSCA FEHYIADW T TVTGLEYRLF EIDSEQLELE FDFRRSGEQP THLGFAVINP LTSDEPPLPQ QWLDDARELL NRQALQAGRQ LSAAEVSQR LAQAGYDFAP YLDHDGELTI GRSGLVLKGR PPVNRHNHYA DNVQLSPYLA TTIDKALYLL LDELGLPQGR VIVRNIERLC CYHTPAGGF SVVLSGIGLN DNELSLSLLV LDEREQICVK LDKVSLYLGK QEVASVDRKH SLLTGEERMA EFKQEAKPQA D DSEAGFGE KILAFIQQEL QDKLGFAADI GESTQVHDLG LDSIMVVQLT DSVNKRFGTK LMPDLFYEKQ QLGELVARLE AA A UniProtKB: RhiE protein, Polyketide synthase domain protein RhiE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 20.0 mM / Component - Name: Tris-HCl / Details: 20 mM Tris-HCl pH 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK III |
| Details | Monodisperse sample in minimal buffer |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-64 (8k x 8k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3579 / Average electron dose: 53.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: JEOL / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X