+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pontibacter korlensis curli subunit CsgA | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | bacterial functional amyloid / PROTEIN FIBRIL | ||||||||||||
| Function / homology | Curlin associated / Curlin associated repeat / pilus / cell adhesion / Curlin associated repeat-containing protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Pontibacter korlensis (bacteria) Pontibacter korlensis (bacteria) | ||||||||||||
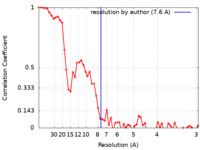
| Method | helical reconstruction / cryo EM / Resolution: 7.6 Å | ||||||||||||
 Authors Authors | Remaut H / Sleutel M / Pradhan B | ||||||||||||
| Funding support |  Belgium, European Union, 3 items Belgium, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural analysis and architectural principles of the bacterial amyloid curli. Authors: Mike Sleutel / Brajabandhu Pradhan / Alexander N Volkov / Han Remaut /  Abstract: Two decades have passed since the initial proposition that amyloids are not only (toxic) byproducts of an unintended aggregation cascade, but that they can also be produced by an organism to serve a ...Two decades have passed since the initial proposition that amyloids are not only (toxic) byproducts of an unintended aggregation cascade, but that they can also be produced by an organism to serve a defined biological function. That revolutionary idea was borne out of the realization that a large fraction of the extracellular matrix that holds Gram-negative cells into a persistent biofilm is composed of protein fibers (curli; tafi) with cross-β architecture, nucleation-dependent polymerization kinetics and classic amyloid tinctorial properties. The list of proteins shown to form so-called functional amyloid fibers in vivo has greatly expanded over the years, but detailed structural insights have not followed at a similar pace in part due to the associated experimental barriers. Here we combine extensive AlphaFold2 modelling and cryo-electron transmission microscopy to propose an atomic model of curli protofibrils, and their higher modes of organization. We uncover an unexpected structural diversity of curli building blocks and fibril architectures. Our results allow for a rationalization of the extreme physico-chemical robustness of curli, as well as earlier observations of inter-species curli promiscuity, and should facilitate further engineering efforts to expand the repertoire of curli-based functional materials. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16431.map.gz emd_16431.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16431-v30.xml emd-16431-v30.xml emd-16431.xml emd-16431.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16431_fsc.xml emd_16431_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_16431.png emd_16431.png | 42 KB | ||
| Filedesc metadata |  emd-16431.cif.gz emd-16431.cif.gz | 5.7 KB | ||
| Others |  emd_16431_half_map_1.map.gz emd_16431_half_map_1.map.gz emd_16431_half_map_2.map.gz emd_16431_half_map_2.map.gz | 9.8 MB 9.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16431 http://ftp.pdbj.org/pub/emdb/structures/EMD-16431 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16431 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16431 | HTTPS FTP |
-Validation report
| Summary document |  emd_16431_validation.pdf.gz emd_16431_validation.pdf.gz | 916 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16431_full_validation.pdf.gz emd_16431_full_validation.pdf.gz | 915.5 KB | Display | |
| Data in XML |  emd_16431_validation.xml.gz emd_16431_validation.xml.gz | 10.7 KB | Display | |
| Data in CIF |  emd_16431_validation.cif.gz emd_16431_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16431 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16431 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16431 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16431 | HTTPS FTP |
-Related structure data
| Related structure data |  8c50MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16431.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16431.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.485 Å | ||||||||||||||||||||||||||||||||||||
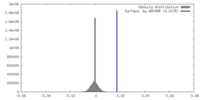
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_16431_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
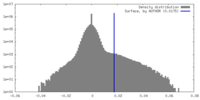
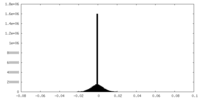
| Density Histograms |
-Half map: #1
| File | emd_16431_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
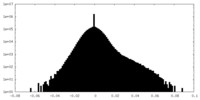
| Density Histograms |
- Sample components
Sample components
-Entire : curli fiber of Pontibacteri korlensis CsgA
| Entire | Name: curli fiber of Pontibacteri korlensis CsgA |
|---|---|
| Components |
|
-Supramolecule #1: curli fiber of Pontibacteri korlensis CsgA
| Supramolecule | Name: curli fiber of Pontibacteri korlensis CsgA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Sample is generated by in vitro polymerisation of Pontibacter korlensis Csga subunits |
|---|---|
| Source (natural) | Organism:  Pontibacter korlensis (bacteria) Pontibacter korlensis (bacteria) |
-Macromolecule #1: Curlin associated repeat-containing protein
| Macromolecule | Name: Curlin associated repeat-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pontibacter korlensis (bacteria) Pontibacter korlensis (bacteria) |
| Molecular weight | Theoretical: 38.033785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QGTTSSATTS QTGNTNTAVI DQVGGLNNSA EASQAGDGNV ATVTQAEGMD NAVYIDQVGL TNTATVLQEG GLDNDADVDQ DGDLNVAYI WQNLGEDNDA DIDQDGTLND AAIEQDGGED NDADIDQDGS ENAAYVGQTG GEDNDADIDQ DGTFNNAYIG Q FGGEDNEA ...String: QGTTSSATTS QTGNTNTAVI DQVGGLNNSA EASQAGDGNV ATVTQAEGMD NAVYIDQVGL TNTATVLQEG GLDNDADVDQ DGDLNVAYI WQNLGEDNDA DIDQDGTLND AAIEQDGGED NDADIDQDGS ENAAYVGQTG GEDNDADIDQ DGTFNNAYIG Q FGGEDNEA DLDQDGDANY AAILQDGGED NDADIDQDGT NNWAETNQIQ GNDNDVEVDQ DGSDNIAEVW QMHGEDNEAN VD QDGDLNN AYILQEGGMN NLADVHQNGV SNTAEIYQYG GMDNEAYLIQ DGDMHTGVIT QSGDGANYAE LNQMGLMNTG SIT QDGMGN SAITSQGGSM NMSTVTQSGT GNISLVNQHG GHHHHHH UniProtKB: Curlin associated repeat-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6 / Details: 15 mM MES pH 6.0 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4455 / Average electron dose: 64.66 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)