+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rosellinia necatrix megabirnavirus 1-W779 full capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | viruses / dsRNA / capsid / cryo-EM / fungus / Megabirnaviridae / mycoviruses / VIRUS | |||||||||
| Function / homology | Coat protein Function and homology information Function and homology information | |||||||||
| Biological species |  Rosellinia necatrix megabirnavirus 1/W779 Rosellinia necatrix megabirnavirus 1/W779 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Wang H / Okamoto K / Miyazaki N / Suzuki N | |||||||||
| Funding support |  Sweden, Sweden,  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2023 Journal: PLoS Pathog / Year: 2023Title: Capsid structure of a fungal dsRNA megabirnavirus reveals its previously unidentified surface architecture. Authors: Han Wang / Lakha Salaipeth / Naoyuki Miyazaki / Nobuhiro Suzuki / Kenta Okamoto /   Abstract: Rosellinia necatrix megabirnavirus 1-W779 (RnMBV1) is a non-enveloped icosahedral double-stranded (ds)RNA virus that infects the ascomycete fungus Rosellinia necatrix, a causative agent that induces ...Rosellinia necatrix megabirnavirus 1-W779 (RnMBV1) is a non-enveloped icosahedral double-stranded (ds)RNA virus that infects the ascomycete fungus Rosellinia necatrix, a causative agent that induces a lethal plant disease white root rot. Herein, we have first resolved the atomic structure of the RnMBV1 capsid at 3.2 Å resolution using cryo-electron microscopy (cryo-EM) single-particle analysis. Compared with other non-enveloped icosahedral dsRNA viruses, the RnMBV1 capsid protein structure exhibits an extra-long C-terminal arm and a surface protrusion domain. In addition, the previously unrecognized crown proteins are identified in a symmetry-expanded cryo-EM model and are present over the 3-fold axes. These exclusive structural features of the RnMBV1 capsid could have been acquired for playing essential roles in transmission and/or particle assembly of the megabirnaviruses. Our findings, therefore, will reinforce the understanding of how the structural and molecular machineries of the megabirnaviruses influence the virulence of the disease-related ascomycete fungus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15855.map.gz emd_15855.map.gz | 236.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15855-v30.xml emd-15855-v30.xml emd-15855.xml emd-15855.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15855.png emd_15855.png | 302.9 KB | ||
| Filedesc metadata |  emd-15855.cif.gz emd-15855.cif.gz | 5.8 KB | ||
| Others |  emd_15855_half_map_1.map.gz emd_15855_half_map_1.map.gz emd_15855_half_map_2.map.gz emd_15855_half_map_2.map.gz | 595.9 MB 595.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15855 http://ftp.pdbj.org/pub/emdb/structures/EMD-15855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15855 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15855 | HTTPS FTP |
-Validation report
| Summary document |  emd_15855_validation.pdf.gz emd_15855_validation.pdf.gz | 928.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15855_full_validation.pdf.gz emd_15855_full_validation.pdf.gz | 927.8 KB | Display | |
| Data in XML |  emd_15855_validation.xml.gz emd_15855_validation.xml.gz | 20.4 KB | Display | |
| Data in CIF |  emd_15855_validation.cif.gz emd_15855_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15855 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15855 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15855 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15855 | HTTPS FTP |
-Related structure data
| Related structure data |  8b4zMC  8b59C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15855.map.gz / Format: CCP4 / Size: 744.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15855.map.gz / Format: CCP4 / Size: 744.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||
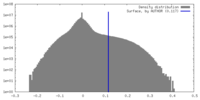
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15855_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
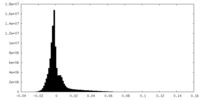
| Density Histograms |
-Half map: #2
| File | emd_15855_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
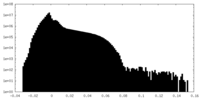
| Density Histograms |
- Sample components
Sample components
-Entire : Rosellinia necatrix megabirnavirus 1/W779
| Entire | Name:  Rosellinia necatrix megabirnavirus 1/W779 Rosellinia necatrix megabirnavirus 1/W779 |
|---|---|
| Components |
|
-Supramolecule #1: Rosellinia necatrix megabirnavirus 1/W779
| Supramolecule | Name: Rosellinia necatrix megabirnavirus 1/W779 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 658904 / Sci species name: Rosellinia necatrix megabirnavirus 1/W779 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein A
| Macromolecule | Name: Major capsid protein A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rosellinia necatrix megabirnavirus 1/W779 / Strain: isolate -/Japan/W779/2001 Rosellinia necatrix megabirnavirus 1/W779 / Strain: isolate -/Japan/W779/2001 |
| Molecular weight | Theoretical: 136.084953 KDa |
| Sequence | String: MSGDNGVYSG SAAYNTATAP KVPVSRATFF QNTKSKDFDF KFADGADAIA NVLQQMEHGV AQHQLGDMNV RTDGLATVSA VLNGRKRKI ANQYMMHFDL FGRAARSTVR MESRIQSFGE GKDVDNFMAK FHNQLSGVYE RRSEGVANFG RILATDTDLG G TSGLSVVF ...String: MSGDNGVYSG SAAYNTATAP KVPVSRATFF QNTKSKDFDF KFADGADAIA NVLQQMEHGV AQHQLGDMNV RTDGLATVSA VLNGRKRKI ANQYMMHFDL FGRAARSTVR MESRIQSFGE GKDVDNFMAK FHNQLSGVYE RRSEGVANFG RILATDTDLG G TSGLSVVF NGLLRGLHHV STVPTPNVAN LPIRNNRDGA GAVVGRGDMP GREFMDSSRI LPPRSSRWYG APGQPIVPPA PN NPPAHVA PMETVMAGLQ KTVMNELNRV IVSIADVPKL PAHRIRNLIA VLAAVSKPNL GFDANRLEDH SCFTKGWLGF NDI LLFPLT VDLFDRVVAN EAGVNDAGFI VPNAAPPQFL QNTNQQVIDF RGVGVGQAGD IPALRLAQSW SDAIGFLLDT IGGE AQLAM GLNDMVAQCF HMHGAQTTML STPIISRADF GVYHNVVTNM YRRLAYMYTR LIRTNAAAGG GAMLDRQHYQ WPTHA KVGF HDDTAVNAAA AAARIHDGLR QPLLDEAFGA GVVQPGNMDL VGAGIDFTRD LTSSLGKAYP EHRPIGADDN KRDLGD FTA GTVDAAASGY EWDNYVYRLF GNMSAMRSKA EFDRLLATFP SSTLSELFIW MGNVGFADTW EERWGYDAAP LCSIPIP AG HDRSMLRNWS WVNVHNVHSV TGTSENVVLA GYVGLSRTHD YIMDTRSTPA TSQGRRLAAM FYYTNADKML SLTFGLAG Q LRAAADTTVA KFQICPHTIA RAQGYIMTDN DPLSDELKGT DFVTEQFSLA GLTNLYLGYF DGLATRLGIY DLRYTYSEY AECRVELHGI QRNFLTDRLD AFVSYKCLHP IMFEYYMCGA NISGGILNGD KAYEQVEMGN IRAYDAMFDT SAARDFNFVG VRGASQQIA AVGGFHIQYK MEVEIQRPGD GTEASRFNVY ERYLNNYLRM SDCAPTSVLN AVSPLFWMAG TTRVVLCEAA N GYKPMAYD ISQTSFWNRE NGLWAFTWGE SEKTHRPNAI PHGTRRLGNS EVLMNSRFSK ILDKKGITKL ETRVGGRKRG DN NDDFVAA DTRMFIIQDV AGGEHAAYSS LRDPGFALVR AAHTWDTFVQ NPRMLLLERG YGNTGFTDTY SAAGIRRTNG HIS LRLSAL TDDFEFTMHP LARAEYKETS RVSLTSMIYV GTAGKDLSLP TGTVEDIIGA VDGMRRVVRT IGGQTIKTAP VVPP TEQRD MVQEERVGTP VKNAGNANPA ADSDNATEGV VEPKN UniProtKB: Coat protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.75 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 12230 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)