+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the SEA complex (Sea2-Sea3 focused map) | |||||||||
 Map data Map data | DeepEMhancer sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
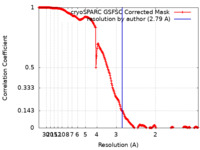
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Tafur L / Loewith R | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Cryo-EM structure of the SEA complex. Authors: Lucas Tafur / Kerstin Hinterndorfer / Caroline Gabus / Chiara Lamanna / Ariane Bergmann / Yashar Sadian / Farzad Hamdi / Fotis L Kyrilis / Panagiotis L Kastritis / Robbie Loewith /   Abstract: The SEA complex (SEAC) is a growth regulator that acts as a GTPase-activating protein (GAP) towards Gtr1, a Rag GTPase that relays nutrient status to the Target of Rapamycin Complex 1 (TORC1) in ...The SEA complex (SEAC) is a growth regulator that acts as a GTPase-activating protein (GAP) towards Gtr1, a Rag GTPase that relays nutrient status to the Target of Rapamycin Complex 1 (TORC1) in yeast. Functionally, the SEAC has been divided into two subcomplexes: SEACIT, which has GAP activity and inhibits TORC1, and SEACAT, which regulates SEACIT. This system is conserved in mammals: the GATOR complex, consisting of GATOR1 (SEACIT) and GATOR2 (SEACAT), transmits amino acid and glucose signals to mTORC1. Despite its importance, the structure of SEAC/GATOR, and thus molecular understanding of its function, is lacking. Here, we solve the cryo-EM structure of the native eight-subunit SEAC. The SEAC has a modular structure in which a COPII-like cage corresponding to SEACAT binds two flexible wings, which correspond to SEACIT. The wings are tethered to the core via Sea3, which forms part of both modules. The GAP mechanism of GATOR1 is conserved in SEACIT, and GAP activity is unaffected by SEACAT in vitro. In vivo, the wings are essential for recruitment of the SEAC to the vacuole, primarily via the EGO complex. Our results indicate that rather than being a direct inhibitor of SEACIT, SEACAT acts as a scaffold for the binding of TORC1 regulators. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15374.map.gz emd_15374.map.gz | 875.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15374-v30.xml emd-15374-v30.xml emd-15374.xml emd-15374.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15374_fsc.xml emd_15374_fsc.xml | 21.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_15374.png emd_15374.png | 34.6 KB | ||
| Masks |  emd_15374_msk_1.map emd_15374_msk_1.map | 1000 MB |  Mask map Mask map | |
| Others |  emd_15374_additional_1.map.gz emd_15374_additional_1.map.gz emd_15374_half_map_1.map.gz emd_15374_half_map_1.map.gz emd_15374_half_map_2.map.gz emd_15374_half_map_2.map.gz | 885.3 MB 928.5 MB 928.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15374 http://ftp.pdbj.org/pub/emdb/structures/EMD-15374 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15374 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15374 | HTTPS FTP |
-Validation report
| Summary document |  emd_15374_validation.pdf.gz emd_15374_validation.pdf.gz | 891.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15374_full_validation.pdf.gz emd_15374_full_validation.pdf.gz | 890.8 KB | Display | |
| Data in XML |  emd_15374_validation.xml.gz emd_15374_validation.xml.gz | 31.3 KB | Display | |
| Data in CIF |  emd_15374_validation.cif.gz emd_15374_validation.cif.gz | 41.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15374 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15374 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15374 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15374 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15374.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15374.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.726 Å | ||||||||||||||||||||||||||||||||||||
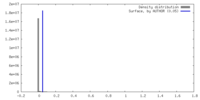
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15374_msk_1.map emd_15374_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
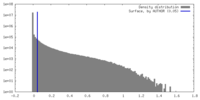
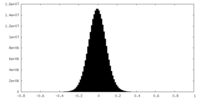
| Density Histograms |
-Additional map: Non-resampled DeepEMhancer sharpened map
| File | emd_15374_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-resampled DeepEMhancer sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
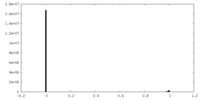
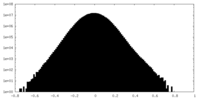
| Density Histograms |
-Half map: #2
| File | emd_15374_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
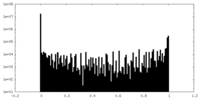
| Density Histograms |
-Half map: #1
| File | emd_15374_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Focused reconstruction on SEAC core subunits Sea2-Seh1 & Sea3-Sec...
| Entire | Name: Focused reconstruction on SEAC core subunits Sea2-Seh1 & Sea3-Sec13, with C2-symmetry expanded particles |
|---|---|
| Components |
|
-Supramolecule #1: Focused reconstruction on SEAC core subunits Sea2-Seh1 & Sea3-Sec...
| Supramolecule | Name: Focused reconstruction on SEAC core subunits Sea2-Seh1 & Sea3-Sec13, with C2-symmetry expanded particles type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#11 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)