[English] 日本語
 Yorodumi
Yorodumi- EMDB-1536: ESCRT-III subunits CHMP2A and CHMP3 assemble in vitro into helica... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1536 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ESCRT-III subunits CHMP2A and CHMP3 assemble in vitro into helical tubular structures | |||||||||
 Map data Map data | ESCRT-III subunits CHMP2A and CHMP3 assemble in vitro into helical tubular structures | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | helical reconstruction | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / negative staining / Resolution: 32.0 Å | |||||||||
 Authors Authors | Lata S / Schoehn G / Jain A / Pires R / Piehler J / Goettlinger H / Weissenhorn W | |||||||||
 Citation Citation |  Journal: Science / Year: 2008 Journal: Science / Year: 2008Title: Helical structures of ESCRT-III are disassembled by VPS4. Authors: Suman Lata / Guy Schoehn / Ankur Jain / Ricardo Pires / Jacob Piehler / Heinrich G Gottlinger / Winfried Weissenhorn /  Abstract: During intracellular membrane trafficking and remodeling, protein complexes known as the ESCRTs (endosomal sorting complexes required for transport) interact with membranes and are required for ...During intracellular membrane trafficking and remodeling, protein complexes known as the ESCRTs (endosomal sorting complexes required for transport) interact with membranes and are required for budding processes directed away from the cytosol, including the budding of intralumenal vesicles to form multivesicular bodies; for the budding of some enveloped viruses; and for daughter cell scission in cytokinesis. We found that the ESCRT-III proteins CHMP2A and CHMP3 (charged multivesicular body proteins 2A and 3) could assemble in vitro into helical tubular structures that expose their membrane interaction sites on the outside of the tubule, whereas the AAA-type adenosine triphosphatase VPS4 could bind on the inside of the tubule and disassemble the tubes upon adenosine triphosphate hydrolysis. CHMP2A and CHMP3 copolymerized in solution, and their membrane targeting was cooperatively enhanced on planar lipid bilayers. Such helical CHMP structures could thus assemble within the neck of an inwardly budding vesicle, catalyzing late steps in budding under the control of VPS4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1536.map.gz emd_1536.map.gz | 8.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1536-v30.xml emd-1536-v30.xml emd-1536.xml emd-1536.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1536.png EMD-1536.png | 263.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1536 http://ftp.pdbj.org/pub/emdb/structures/EMD-1536 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1536 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1536 | HTTPS FTP |
-Validation report
| Summary document |  emd_1536_validation.pdf.gz emd_1536_validation.pdf.gz | 232.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1536_full_validation.pdf.gz emd_1536_full_validation.pdf.gz | 231.2 KB | Display | |
| Data in XML |  emd_1536_validation.xml.gz emd_1536_validation.xml.gz | 5.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1536 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1536 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1536 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1536 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1536.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1536.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ESCRT-III subunits CHMP2A and CHMP3 assemble in vitro into helical tubular structures | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
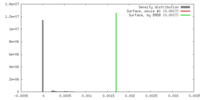
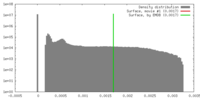
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CHMP proteins
| Entire | Name: CHMP proteins |
|---|---|
| Components |
|
-Supramolecule #1000: CHMP proteins
| Supramolecule | Name: CHMP proteins / type: sample / ID: 1000 / Oligomeric state: oligomer of dimers / Number unique components: 1 |
|---|
-Macromolecule #1: CHMP
| Macromolecule | Name: CHMP / type: protein_or_peptide / ID: 1 / Name.synonym: CHMP Details: CHMP2AdeltaC (9-161)-CHMP3deltaC(9 to 183) . MBP attached to CHMP2AdeltaC Oligomeric state: oligomer of dimers / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 20 mM HEPES, pH 7.6, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: Vitrified samples were prepared according to the method of Dubochet et al., |
| Grid | Details: Quantifoil R2/2 |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 100 K / Instrument: OTHER / Details: Vitrification instrument: Zeiss / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200T |
|---|---|
| Temperature | Average: 100 K |
| Alignment procedure | Legacy - Astigmatism: orrected at 100,000 times magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 10 / Average electron dose: 10 e/Å2 / Details: Images binned after scanning / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 39500 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | Particles were selected by hand using X3d |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 32 Å Applied symmetry - Helical parameters - Δ&Phi: 21.71 ° Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 32.0 Å / Resolution method: OTHER / Software - Name: IHRSR / Details: 2500 out of 4400 particles have been used |
| CTF correction | Details: each particle |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera



 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)