+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | IHMof beta-cardiac myosin heads region | ||||||||||||
 Map data Map data | map of the IHMof beta-cardiac myosin heads region | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Cardiac Myosin / Myosin / Human / folded-back off state / head-head region / CONTRACTILE PROTEIN | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
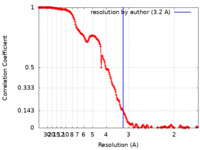
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Grinzato A / Kandiah E / Robert-Paganin J | ||||||||||||
| Funding support |  United States, United States,  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structure of the folded-back state of human β-cardiac myosin. Authors: Alessandro Grinzato / Daniel Auguin / Carlos Kikuti / Neha Nandwani / Dihia Moussaoui / Divya Pathak / Eaazhisai Kandiah / Kathleen M Ruppel / James A Spudich / Anne Houdusse / Julien Robert-Paganin /   Abstract: To save energy and precisely regulate cardiac contractility, cardiac muscle myosin heads are sequestered in an 'off' state that can be converted to an 'on' state when exertion is increased. The 'off' ...To save energy and precisely regulate cardiac contractility, cardiac muscle myosin heads are sequestered in an 'off' state that can be converted to an 'on' state when exertion is increased. The 'off' state is equated with a folded-back structure known as the interacting-heads motif (IHM), which is a regulatory feature of all class-2 muscle and non-muscle myosins. We report here the human β-cardiac myosin IHM structure determined by cryo-electron microscopy to 3.6 Å resolution, providing details of all the interfaces stabilizing the 'off' state. The structure shows that these interfaces are hot spots of hypertrophic cardiomyopathy mutations that are thought to cause hypercontractility by destabilizing the 'off' state. Importantly, the cardiac and smooth muscle myosin IHM structures dramatically differ, providing structural evidence for the divergent physiological regulation of these muscle types. The cardiac IHM structure will facilitate development of clinically useful new molecules that modulate IHM stability. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15354.map.gz emd_15354.map.gz | 450.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15354-v30.xml emd-15354-v30.xml emd-15354.xml emd-15354.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15354_fsc.xml emd_15354_fsc.xml | 17.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_15354.png emd_15354.png | 100.9 KB | ||
| Masks |  emd_15354_msk_1.map emd_15354_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Others |  emd_15354_half_map_1.map.gz emd_15354_half_map_1.map.gz emd_15354_half_map_2.map.gz emd_15354_half_map_2.map.gz | 442.3 MB 442.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15354 http://ftp.pdbj.org/pub/emdb/structures/EMD-15354 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15354 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15354 | HTTPS FTP |
-Validation report
| Summary document |  emd_15354_validation.pdf.gz emd_15354_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15354_full_validation.pdf.gz emd_15354_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_15354_validation.xml.gz emd_15354_validation.xml.gz | 24.9 KB | Display | |
| Data in CIF |  emd_15354_validation.cif.gz emd_15354_validation.cif.gz | 32.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15354 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15354 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15354 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15354 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15354.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15354.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of the IHMof beta-cardiac myosin heads region | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
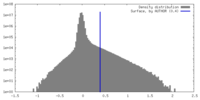
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15354_msk_1.map emd_15354_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
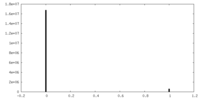
| Density Histograms |
-Half map: half map of the IHMof beta-cardiac myosin heads region
| File | emd_15354_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of the IHMof beta-cardiac myosin heads region | ||||||||||||
| Projections & Slices |
| ||||||||||||
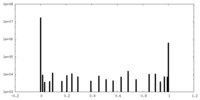
| Density Histograms |
-Half map: half map of the IHMof beta-cardiac myosin heads region
| File | emd_15354_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of the IHMof beta-cardiac myosin heads region | ||||||||||||
| Projections & Slices |
| ||||||||||||
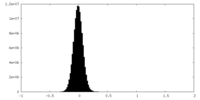
| Density Histograms |
- Sample components
Sample components
-Entire : human beta-cardiac myosin folded-back off state
| Entire | Name: human beta-cardiac myosin folded-back off state |
|---|---|
| Components |
|
-Supramolecule #1: human beta-cardiac myosin folded-back off state
| Supramolecule | Name: human beta-cardiac myosin folded-back off state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 8.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)