+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | H1-bound palindromic nucleosome, state 2 | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.0 Å | ||||||||||||||||||||||||
 Authors Authors | Alegrio Louro J / Beinsteiner B / Cheng TC / Patel AKM / Boopathi R / Angelov D / Hamiche A / Bednar J / Kale S / Dimitrov S / Klaholz B | ||||||||||||||||||||||||
| Funding support |  France, European Union, 7 items France, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Nucleosome dyad determines the H1 C-terminus collapse on distinct DNA arms. Authors: Jaime Alegrio Louro / Ramachandran Boopathi / Brice Beinsteiner / Abdul Kareem Mohideen Patel / Tat Cheung Cheng / Dimitar Angelov / Ali Hamiche / Jan Bendar / Seyit Kale / Bruno P Klaholz / Stefan Dimitrov /   Abstract: Nucleosomes are symmetric structures. However, binding of linker histones generates an inherently asymmetric H1-nucleosome complex, and whether this asymmetry is transmitted to the overall nucleosome ...Nucleosomes are symmetric structures. However, binding of linker histones generates an inherently asymmetric H1-nucleosome complex, and whether this asymmetry is transmitted to the overall nucleosome structure, and therefore also to chromatin, is unclear. Efforts to investigate potential asymmetry due to H1s have been hampered by the DNA sequence, which naturally differs in each gyre. To overcome this issue, we designed and analyzed by cryo-EM a nucleosome reconstituted with a palindromic (601L) 197-bp DNA. As in the non-palindromic 601 sequence, H1 restricts linker DNA flexibility but reveals partial asymmetrical unwrapping. However, in contrast to the non-palindromic nucleosome, in the palindromic nucleosome H1 CTD collapses to the proximal linker. Molecular dynamics simulations show that this could be dictated by a slightly tilted orientation of the globular domain (GD) of H1, which could be linked to the DNA sequence of the nucleosome dyad. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15146.map.gz emd_15146.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15146-v30.xml emd-15146-v30.xml emd-15146.xml emd-15146.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15146.png emd_15146.png | 92.5 KB | ||
| Others |  emd_15146_half_map_1.map.gz emd_15146_half_map_1.map.gz emd_15146_half_map_2.map.gz emd_15146_half_map_2.map.gz | 31.4 MB 31.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15146 http://ftp.pdbj.org/pub/emdb/structures/EMD-15146 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15146 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15146 | HTTPS FTP |
-Validation report
| Summary document |  emd_15146_validation.pdf.gz emd_15146_validation.pdf.gz | 660.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15146_full_validation.pdf.gz emd_15146_full_validation.pdf.gz | 659.7 KB | Display | |
| Data in XML |  emd_15146_validation.xml.gz emd_15146_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_15146_validation.cif.gz emd_15146_validation.cif.gz | 13.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15146 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15146 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15146 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15146 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15146.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15146.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||
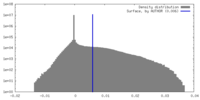
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15146_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
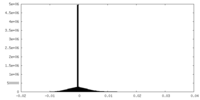
| Density Histograms |
-Half map: #2
| File | emd_15146_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 197-bp H1-bound palindromic nucleosome
| Entire | Name: 197-bp H1-bound palindromic nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: 197-bp H1-bound palindromic nucleosome
| Supramolecule | Name: 197-bp H1-bound palindromic nucleosome / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#10 Details: 601L nucleosome with 25-bp sequence symmetric linkers and bound to linker histone H1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120 KDa |
-Supramolecule #2: Histone H1.0-B
| Supramolecule | Name: Histone H1.0-B / type: complex / ID: 2 / Chimera: Yes / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism: |
-Supramolecule #3: Core histone octamer
| Supramolecule | Name: Core histone octamer / type: complex / ID: 3 / Chimera: Yes / Parent: 1 / Macromolecule list: #4-#10 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: SUPER-RESOLUTION / #0 - Number grids imaged: 1 / #0 - Number real images: 3999 / #0 - Average exposure time: 10.0 sec. / #0 - Average electron dose: 49.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Detector mode: SUPER-RESOLUTION / #1 - Number grids imaged: 1 / #1 - Number real images: 4891 / #1 - Average exposure time: 5.5 sec. / #1 - Average electron dose: 40.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)