[English] 日本語
 Yorodumi
Yorodumi- EMDB-15136: In-cell structure of the actin filament Arp2/3 complex branch jun... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In-cell structure of the actin filament Arp2/3 complex branch junction in lamellipodia of WT B16-F1 mouse melanoma cells | |||||||||
 Map data Map data | Structure of the Arp2/3 actin branch junction as found in B16-F1 mouse melanoma cell lamellipodia | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
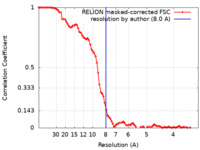
| Method | subtomogram averaging / cryo EM / Resolution: 8.0 Å | |||||||||
 Authors Authors | Faessler F / Javoor MG / Datler J / Doering H / Hofer FW / Dimchev G / Hodirnau VV / Rottner K / Schur FKM | |||||||||
| Funding support |  Austria, 1 items Austria, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: ArpC5 isoforms regulate Arp2/3 complex-dependent protrusion through differential Ena/VASP positioning. Authors: Florian Fäßler / Manjunath G Javoor / Julia Datler / Hermann Döring / Florian W Hofer / Georgi Dimchev / Victor-Valentin Hodirnau / Jan Faix / Klemens Rottner / Florian K M Schur /   Abstract: Regulation of the Arp2/3 complex is required for productive nucleation of branched actin networks. An emerging aspect of regulation is the incorporation of subunit isoforms into the Arp2/3 complex. ...Regulation of the Arp2/3 complex is required for productive nucleation of branched actin networks. An emerging aspect of regulation is the incorporation of subunit isoforms into the Arp2/3 complex. Specifically, both ArpC5 subunit isoforms, ArpC5 and ArpC5L, have been reported to fine-tune nucleation activity and branch junction stability. We have combined reverse genetics and cellular structural biology to describe how ArpC5 and ArpC5L differentially affect cell migration. Both define the structural stability of ArpC1 in branch junctions and, in turn, by determining protrusion characteristics, affect protein dynamics and actin network ultrastructure. ArpC5 isoforms also affect the positioning of members of the Ena/Vasodilator-stimulated phosphoprotein (VASP) family of actin filament elongators, which mediate ArpC5 isoform-specific effects on the actin assembly level. Our results suggest that ArpC5 and Ena/VASP proteins are part of a signaling pathway enhancing cell migration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15136.map.gz emd_15136.map.gz | 49.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15136-v30.xml emd-15136-v30.xml emd-15136.xml emd-15136.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15136_fsc.xml emd_15136_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_15136.png emd_15136.png | 43.8 KB | ||
| Masks |  emd_15136_msk_1.map emd_15136_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Others |  emd_15136_half_map_1.map.gz emd_15136_half_map_1.map.gz emd_15136_half_map_2.map.gz emd_15136_half_map_2.map.gz | 27 MB 27 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15136 http://ftp.pdbj.org/pub/emdb/structures/EMD-15136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15136 | HTTPS FTP |
-Validation report
| Summary document |  emd_15136_validation.pdf.gz emd_15136_validation.pdf.gz | 844.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15136_full_validation.pdf.gz emd_15136_full_validation.pdf.gz | 844.2 KB | Display | |
| Data in XML |  emd_15136_validation.xml.gz emd_15136_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_15136_validation.cif.gz emd_15136_validation.cif.gz | 20 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15136 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15136 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15136 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15136 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15136.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15136.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the Arp2/3 actin branch junction as found in B16-F1 mouse melanoma cell lamellipodia | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.693 Å | ||||||||||||||||||||||||||||||||||||
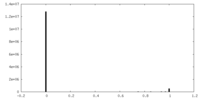
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15136_msk_1.map emd_15136_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
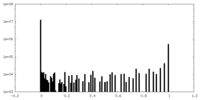
| Density Histograms |
-Half map: Halfmap 1 after subtomogram averaging and multi-particle refinemnt
| File | emd_15136_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 1 after subtomogram averaging and multi-particle refinemnt | ||||||||||||
| Projections & Slices |
| ||||||||||||
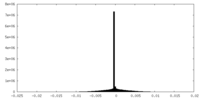
| Density Histograms |
-Half map: Halfmap 1 after subtomogram averaging and multi-particle refinemnt
| File | emd_15136_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 1 after subtomogram averaging and multi-particle refinemnt | ||||||||||||
| Projections & Slices |
| ||||||||||||
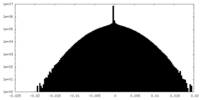
| Density Histograms |
- Sample components
Sample components
-Entire : In-cell structure of the actin filament Arp2/3 complex branch jun...
| Entire | Name: In-cell structure of the actin filament Arp2/3 complex branch junction in lamellipodia of WT B16-F1 mouse melanoma cells |
|---|---|
| Components |
|
-Supramolecule #1: In-cell structure of the actin filament Arp2/3 complex branch jun...
| Supramolecule | Name: In-cell structure of the actin filament Arp2/3 complex branch junction in lamellipodia of WT B16-F1 mouse melanoma cells type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 Component:
Details: Adjust to pH 6.2 using NaOH Immediately prior to use, add Phalloidin to a final concentration of 1ug/ml | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR Details: The grids were coated with 25ug/ml laminin for 1 hour prior to seeding the cells | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 2.79 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)