[English] 日本語
 Yorodumi
Yorodumi- EMDB-15024: Structure of the human sodium/bile acid cotransporter (NTCP) in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody | |||||||||
 Map data Map data | Nanodisc-reconstituted human NTCP in a complex with NTCP_Fab12 and a nanobody in the presence of substrate. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationbile acid:sodium symporter activity / regulation of bile acid secretion / bile acid and bile salt transport / bile acid signaling pathway / Recycling of bile acids and salts / response to nutrient levels / response to organic cyclic compound / response to estrogen / cellular response to xenobiotic stimulus / virus receptor activity ...bile acid:sodium symporter activity / regulation of bile acid secretion / bile acid and bile salt transport / bile acid signaling pathway / Recycling of bile acids and salts / response to nutrient levels / response to organic cyclic compound / response to estrogen / cellular response to xenobiotic stimulus / virus receptor activity / basolateral plasma membrane / response to ethanol / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
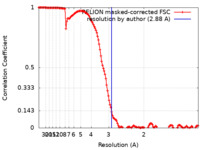
| Method | single particle reconstruction / cryo EM / Resolution: 2.88 Å | |||||||||
 Authors Authors | Liu H / Irobalieva RN / Bang-Sorensen R / Nosol K / Mukherjee S / Agrawal P / Stieger B / Kossiakoff AA / Locher KP | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2022 Journal: Cell Res / Year: 2022Title: Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Authors: Hongtao Liu / Rossitza N Irobalieva / Rose Bang-Sørensen / Kamil Nosol / Somnath Mukherjee / Parth Agrawal / Bruno Stieger / Anthony A Kossiakoff / Kaspar P Locher /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15024.map.gz emd_15024.map.gz | 201.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15024-v30.xml emd-15024-v30.xml emd-15024.xml emd-15024.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15024_fsc.xml emd_15024_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15024.png emd_15024.png | 91.5 KB | ||
| Others |  emd_15024_half_map_1.map.gz emd_15024_half_map_1.map.gz emd_15024_half_map_2.map.gz emd_15024_half_map_2.map.gz | 171.4 MB 171.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15024 http://ftp.pdbj.org/pub/emdb/structures/EMD-15024 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15024 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15024 | HTTPS FTP |
-Validation report
| Summary document |  emd_15024_validation.pdf.gz emd_15024_validation.pdf.gz | 824.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15024_full_validation.pdf.gz emd_15024_full_validation.pdf.gz | 824.2 KB | Display | |
| Data in XML |  emd_15024_validation.xml.gz emd_15024_validation.xml.gz | 20.9 KB | Display | |
| Data in CIF |  emd_15024_validation.cif.gz emd_15024_validation.cif.gz | 27.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15024 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15024 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15024 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15024 | HTTPS FTP |
-Related structure data
| Related structure data |  7zyiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15024.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15024.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nanodisc-reconstituted human NTCP in a complex with NTCP_Fab12 and a nanobody in the presence of substrate. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
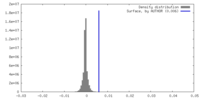
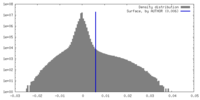
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15024_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
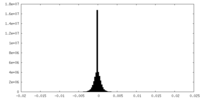
| Density Histograms |
-Half map: #1
| File | emd_15024_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
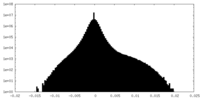
| Density Histograms |
- Sample components
Sample components
-Entire : sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody
| Entire | Name: sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody |
|---|---|
| Components |
|
-Supramolecule #1: sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody
| Supramolecule | Name: sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: Sodium/bile acid cotransporter
| Macromolecule | Name: Sodium/bile acid cotransporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.149949 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEAHNASAPF NFTLPPNFGK RPTDLALSVI LVFMLFFIML SLGCTMEFSK IKAHLWKPKG LAIALVAQYG IMPLTAFVLG KVFRLKNIE ALAILVCGCS PGGNLSNVFS LAMKGDMNLS IVMTTCSTFC ALGMMPLLLY IYSRGIYDGD LKDKVPYKGI V ISLVLVLI ...String: MEAHNASAPF NFTLPPNFGK RPTDLALSVI LVFMLFFIML SLGCTMEFSK IKAHLWKPKG LAIALVAQYG IMPLTAFVLG KVFRLKNIE ALAILVCGCS PGGNLSNVFS LAMKGDMNLS IVMTTCSTFC ALGMMPLLLY IYSRGIYDGD LKDKVPYKGI V ISLVLVLI PCTIGIVLKS KRPQYMRYVI KGGMIIILLC SVAVTVLSAI NVGKSIMFAM TPLLIATSSL MPFIGFLLGY VL SALFCLN GRCRRTVSME TGCQNVQLCS TILNVAFPPE VIGPLFFFPL LYMIFQLGEG LLLIAIFWCY EKFKTPKDKT KMI YTAATT EETIPGALGN GTYKGEDCSP CTA |
-Macromolecule #2: heavy chain of Fab
| Macromolecule | Name: heavy chain of Fab / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.582521 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVSYSSIHWV RQAPGKGLEW VASISSSYGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYMKQQSQM WYQRYWGFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVSYSSIHWV RQAPGKGLEW VASISSSYGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYMKQQSQM WYQRYWGFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCDKTHT |
-Macromolecule #3: light chain of Fab
| Macromolecule | Name: light chain of Fab / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.417971 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYWSPITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYWSPITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: Nanobody
| Macromolecule | Name: Nanobody / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.118386 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLQESGGGL VQPGGSLRLS CAASGRTISR YAMSWFRQAP GKEREFVAVA RRSGDGAFYA DSVQGRFTVS RDDAKNTVYL QMNSLKPED TAVYYCAIDS DTFYSGSYDY WGQGTQVTVS S |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #6: GLYCOCHENODEOXYCHOLIC ACID
| Macromolecule | Name: GLYCOCHENODEOXYCHOLIC ACID / type: ligand / ID: 6 / Number of copies: 2 / Formula: CHO |
|---|---|
| Molecular weight | Theoretical: 449.623 Da |
| Chemical component information |  ChemComp-CHO: |
-Macromolecule #7: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 7 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 13208 / Average exposure time: 2.0 sec. / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)