[English] 日本語
 Yorodumi
Yorodumi- EMDB-14968: HOPS tethering complex from yeast, local refinement map of the ba... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HOPS tethering complex from yeast, local refinement map of the backbone part of the complex | ||||||||||||
 Map data Map data | Tethering complex HOPS, local refinement map of the backbone part of the complex | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | ||||||||||||
 Authors Authors | Shvarev D / Schoppe J / Koenig C / Perz A / Fuellbrunn N / Kiontke S / Langemeyer L / Januliene D / Schnelle K / Kuemmel D ...Shvarev D / Schoppe J / Koenig C / Perz A / Fuellbrunn N / Kiontke S / Langemeyer L / Januliene D / Schnelle K / Kuemmel D / Froehlich F / Moeller A / Ungermann C | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery. Authors: Dmitry Shvarev / Jannis Schoppe / Caroline König / Angela Perz / Nadia Füllbrunn / Stephan Kiontke / Lars Langemeyer / Dovile Januliene / Kilian Schnelle / Daniel Kümmel / Florian ...Authors: Dmitry Shvarev / Jannis Schoppe / Caroline König / Angela Perz / Nadia Füllbrunn / Stephan Kiontke / Lars Langemeyer / Dovile Januliene / Kilian Schnelle / Daniel Kümmel / Florian Fröhlich / Arne Moeller / Christian Ungermann /  Abstract: Lysosomes are essential for cellular recycling, nutrient signaling, autophagy, and pathogenic bacteria and viruses invasion. Lysosomal fusion is fundamental to cell survival and requires HOPS, a ...Lysosomes are essential for cellular recycling, nutrient signaling, autophagy, and pathogenic bacteria and viruses invasion. Lysosomal fusion is fundamental to cell survival and requires HOPS, a conserved heterohexameric tethering complex. On the membranes to be fused, HOPS binds small membrane-associated GTPases and assembles SNAREs for fusion, but how the complex fulfills its function remained speculative. Here, we used cryo-electron microscopy to reveal the structure of HOPS. Unlike previously reported, significant flexibility of HOPS is confined to its extremities, where GTPase binding occurs. The SNARE-binding module is firmly attached to the core, therefore, ideally positioned between the membranes to catalyze fusion. Our data suggest a model for how HOPS fulfills its dual functionality of tethering and fusion and indicate why it is an essential part of the membrane fusion machinery. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14968.map.gz emd_14968.map.gz | 1.1 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14968-v30.xml emd-14968-v30.xml emd-14968.xml emd-14968.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14968_fsc.xml emd_14968_fsc.xml | 22.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_14968.png emd_14968.png | 48 KB | ||
| Others |  emd_14968_half_map_1.map.gz emd_14968_half_map_1.map.gz emd_14968_half_map_2.map.gz emd_14968_half_map_2.map.gz | 1.1 GB 1.1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14968 http://ftp.pdbj.org/pub/emdb/structures/EMD-14968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14968 | HTTPS FTP |
-Validation report
| Summary document |  emd_14968_validation.pdf.gz emd_14968_validation.pdf.gz | 986 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14968_full_validation.pdf.gz emd_14968_full_validation.pdf.gz | 985.5 KB | Display | |
| Data in XML |  emd_14968_validation.xml.gz emd_14968_validation.xml.gz | 32.2 KB | Display | |
| Data in CIF |  emd_14968_validation.cif.gz emd_14968_validation.cif.gz | 42.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14968 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14968 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14968 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14968 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14968.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14968.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tethering complex HOPS, local refinement map of the backbone part of the complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.924 Å | ||||||||||||||||||||||||||||||||||||
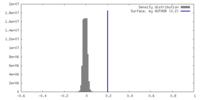
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Tethering complex HOPS, local refinement map of the...
| File | emd_14968_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tethering complex HOPS, local refinement map of the backbone part of the complex - half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
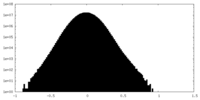
| Density Histograms |
-Half map: Tethering complex HOPS, local refinement map of the...
| File | emd_14968_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tethering complex HOPS, local refinement map of the backbone part of the complex - half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
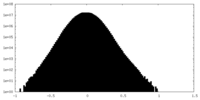
| Density Histograms |
- Sample components
Sample components
-Entire : Tethering complex HOPS
| Entire | Name: Tethering complex HOPS |
|---|---|
| Components |
|
-Supramolecule #1: Tethering complex HOPS
| Supramolecule | Name: Tethering complex HOPS / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)