[English] 日本語
 Yorodumi
Yorodumi- EMDB-14819: Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Re... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state, MD focused map Respiratory complex I Respiratory complex I | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
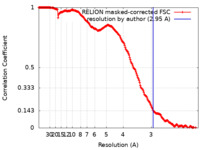
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.95 Å cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Kravchuk V / Kampjut D / Sazanov L | |||||||||
| Funding support |  Austria, 1 items Austria, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: A universal coupling mechanism of respiratory complex I. Authors: Vladyslav Kravchuk / Olga Petrova / Domen Kampjut / Anna Wojciechowska-Bason / Zara Breese / Leonid Sazanov /    Abstract: Complex I is the first enzyme in the respiratory chain, which is responsible for energy production in mitochondria and bacteria. Complex I couples the transfer of two electrons from NADH to quinone ...Complex I is the first enzyme in the respiratory chain, which is responsible for energy production in mitochondria and bacteria. Complex I couples the transfer of two electrons from NADH to quinone and the translocation of four protons across the membrane, but the coupling mechanism remains contentious. Here we present cryo-electron microscopy structures of Escherichia coli complex I (EcCI) in different redox states, including catalytic turnover. EcCI exists mostly in the open state, in which the quinone cavity is exposed to the cytosol, allowing access for water molecules, which enable quinone movements. Unlike the mammalian paralogues, EcCI can convert to the closed state only during turnover, showing that closed and open states are genuine turnover intermediates. The open-to-closed transition results in the tightly engulfed quinone cavity being connected to the central axis of the membrane arm, a source of substrate protons. Consistently, the proportion of the closed state increases with increasing pH. We propose a detailed but straightforward and robust mechanism comprising a 'domino effect' series of proton transfers and electrostatic interactions: the forward wave ('dominoes stacking') primes the pump, and the reverse wave ('dominoes falling') results in the ejection of all pumped protons from the distal subunit NuoL. This mechanism explains why protons exit exclusively from the NuoL subunit and is supported by our mutagenesis data. We contend that this is a universal coupling mechanism of complex I and related enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14819.map.gz emd_14819.map.gz | 274.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14819-v30.xml emd-14819-v30.xml emd-14819.xml emd-14819.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14819_fsc.xml emd_14819_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14819.png emd_14819.png | 64.3 KB | ||
| Others |  emd_14819_half_map_1.map.gz emd_14819_half_map_1.map.gz emd_14819_half_map_2.map.gz emd_14819_half_map_2.map.gz | 391.1 MB 391.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14819 http://ftp.pdbj.org/pub/emdb/structures/EMD-14819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14819 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14819 | HTTPS FTP |
-Related structure data
| Related structure data |  7p61C  7p62C  7p63C  7p64C  7p69C  7p7cC  7p7eC  7p7jC  7p7kC  7p7lC  7p7mC  7z7rC  7z7sC  7z7tC  7z7vC  7z80C  7z83C  7z84C  7zc5C  7zciC  7zd6C  7zdhC  7zdjC  7zdmC  7zdpC  7zebC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14819.map.gz / Format: CCP4 / Size: 488.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14819.map.gz / Format: CCP4 / Size: 488.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_14819_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
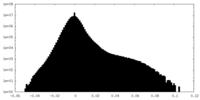
| Projections & Slices |
| ||||||||||||
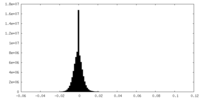
| Density Histograms |
-Half map: #2
| File | emd_14819_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
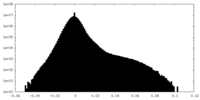
| Projections & Slices |
| ||||||||||||
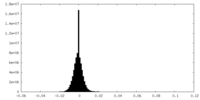
| Density Histograms |
- Sample components
Sample components
-Entire : Complex I
| Entire | Name: Complex I Respiratory complex I Respiratory complex I |
|---|---|
| Components |
|
-Supramolecule #1: Complex I
| Supramolecule | Name: Complex I / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#13 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) / Strain: MC4100 Escherichia coli (E. coli) / Strain: MC4100 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.9 nm | |||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Sampling interval: 5.0 µm / Number grids imaged: 1 / Number real images: 11316 / Average electron dose: 80.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











































































 Z
Z Y
Y X
X