[English] 日本語
 Yorodumi
Yorodumi- EMDB-14576: CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H4 Q98R H100E nanobod... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H4 Q98R H100E nanobody complex in 2Up1Down conformation | |||||||||
 Map data Map data | Sharpened map, output from postprocessing. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information virion component / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ... virion component / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ... virion component / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / virion component / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 / Severe acute respiratory syndrome coronavirus 2 /   Lama glama (llama) / Lama glama (llama) /   Escherichia virus T4 Escherichia virus T4 | |||||||||
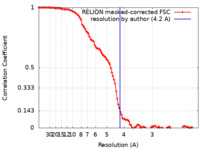
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.2 Å cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Weckener M / Naismith JH | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes. Authors: Halina Mikolajek / Miriam Weckener / Z Faidon Brotzakis / Jiandong Huo / Evmorfia V Dalietou / Audrey Le Bas / Pietro Sormanni / Peter J Harrison / Philip N Ward / Steven Truong / Lucile ...Authors: Halina Mikolajek / Miriam Weckener / Z Faidon Brotzakis / Jiandong Huo / Evmorfia V Dalietou / Audrey Le Bas / Pietro Sormanni / Peter J Harrison / Philip N Ward / Steven Truong / Lucile Moynie / Daniel K Clare / Maud Dumoux / Joshua Dormon / Chelsea Norman / Naveed Hussain / Vinod Vogirala / Raymond J Owens / Michele Vendruscolo / James H Naismith /   Abstract: Camelid single-domain antibodies, also known as nanobodies, can be readily isolated from naïve libraries for specific targets but often bind too weakly to their targets to be immediately useful. ...Camelid single-domain antibodies, also known as nanobodies, can be readily isolated from naïve libraries for specific targets but often bind too weakly to their targets to be immediately useful. Laboratory-based genetic engineering methods to enhance their affinity, termed maturation, can deliver useful reagents for different areas of biology and potentially medicine. Using the receptor binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein and a naïve library, we generated closely related nanobodies with micromolar to nanomolar binding affinities. By analyzing the structure-activity relationship using X-ray crystallography, cryoelectron microscopy, and biophysical methods, we observed that higher conformational entropy losses in the formation of the spike protein-nanobody complex are associated with tighter binding. To investigate this, we generated structural ensembles of the different complexes from electron microscopy maps and correlated the conformational fluctuations with binding affinity. This insight guided the engineering of a nanobody with improved affinity for the spike protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14576.map.gz emd_14576.map.gz | 10.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14576-v30.xml emd-14576-v30.xml emd-14576.xml emd-14576.xml | 29.2 KB 29.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14576_fsc.xml emd_14576_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14576.png emd_14576.png | 121 KB | ||
| Masks |  emd_14576_msk_1.map emd_14576_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_14576_additional_1.map.gz emd_14576_additional_1.map.gz emd_14576_additional_2.map.gz emd_14576_additional_2.map.gz emd_14576_half_map_1.map.gz emd_14576_half_map_1.map.gz emd_14576_half_map_2.map.gz emd_14576_half_map_2.map.gz | 80.8 MB 5.2 MB 80.8 MB 81.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14576 http://ftp.pdbj.org/pub/emdb/structures/EMD-14576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14576 | HTTPS FTP |
-Related structure data
| Related structure data |  7z9rMC  7z1aC  7z1bC  7z1cC  7z1dC  7z1eC  7z6vC  7z7xC  7z85C  7z86C  7z9qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14576.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14576.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map, output from postprocessing. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||
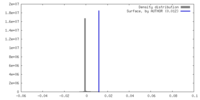
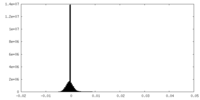
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14576_msk_1.map emd_14576_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
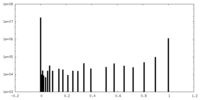
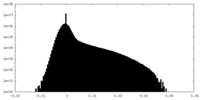
| Density Histograms |
-Additional map: Map from final 3D refinement.
| File | emd_14576_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from final 3D refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
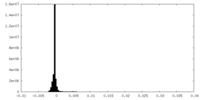
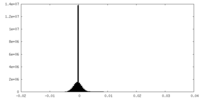
| Density Histograms |
-Additional map: Sharpened map, output from LocScale.
| File | emd_14576_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map, output from LocScale. | ||||||||||||
| Projections & Slices |
| ||||||||||||
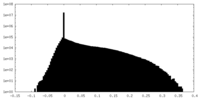
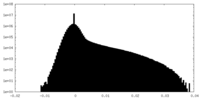
| Density Histograms |
-Half map: Half map from final 3D refinement.
| File | emd_14576_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from final 3D refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map from final 3D refinement.
| File | emd_14576_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from final 3D refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between Spike and nanobody H11-H4 Q98R H100E
| Entire | Name: Complex between Spike and nanobody H11-H4 Q98R H100E |
|---|---|
| Components |
|
-Supramolecule #1: Complex between Spike and nanobody H11-H4 Q98R H100E
| Supramolecule | Name: Complex between Spike and nanobody H11-H4 Q98R H100E / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The Spike protein is found in the 2Up1Down conformation. |
|---|---|
| Molecular weight | Theoretical: 15 KDa |
-Supramolecule #2: Spike glycoprotein trimer with fibritin tag
| Supramolecule | Name: Spike glycoprotein trimer with fibritin tag / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant strain: HEK 293T Homo sapiens (human) / Recombinant strain: HEK 293T |
-Supramolecule #3: Nanobody H11-H4 Q98R H100E
| Supramolecule | Name: Nanobody H11-H4 Q98R H100E / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant strain: WK6 Escherichia coli (E. coli) / Recombinant strain: WK6 |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia virus T4 Escherichia virus T4 |
| Molecular weight | Theoretical: 139.877906 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGAALQIPFA MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STASALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQGSGYIPE APRDGQAYVR KDGEWVLLST FLSLLNDIFE AQKIEWHEKH HHHHH |
-Macromolecule #2: Nanobody H11-H4 Q98R H100E
| Macromolecule | Name: Nanobody H11-H4 Q98R H100E / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Molecular weight | Theoretical: 14.107723 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: QVQLVESGGG LMQAGGSLRL SCAVSGRTFS TAAMGWFRQA PGKEREFVAA IRWSGGSAYY ADSVKGRFTI SRDKAKNTVY LQMNSLKYE DTAVYYCART EYVSYLLSDY ATWPYDYWGQ GTQVTVSS |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 26 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: Grids were glow discharged twice, 60s each. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 289.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Details | EPU auto-function coma free correction |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Average exposure time: 3.85 sec. / Average electron dose: 50.0 e/Å2 / Details: The images were collected as 50 frame movies |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)