+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Leishmania RNA virus 1 virion | |||||||||
 マップデータ マップデータ | Virion structure | |||||||||
 試料 試料 |
| |||||||||
| 機能・相同性 | Totivirus coat / Totivirus coat protein / Capsid protein 機能・相同性情報 機能・相同性情報 | |||||||||
| 生物種 |  Leishmania RNA virus 1 - 4 (ウイルス) Leishmania RNA virus 1 - 4 (ウイルス) | |||||||||
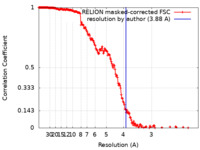
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.88 Å | |||||||||
 データ登録者 データ登録者 | Prochazkova M / Plevka P | |||||||||
| 資金援助 |  チェコ, 1件 チェコ, 1件
| |||||||||
 引用 引用 |  ジャーナル: Virology / 年: 2022 ジャーナル: Virology / 年: 2022タイトル: Virion structure of Leishmania RNA virus 1. 著者: Michaela Procházková / Tibor Füzik / Danyil Grybchuk / Vyacheslav Yurchenko / Pavel Plevka /  要旨: The presence of Leishmania RNA virus 1 (LRV1) enables Leishmania protozoan parasites to cause more severe disease than the virus-free strains. The structure of LRV1 virus-like particles has been ...The presence of Leishmania RNA virus 1 (LRV1) enables Leishmania protozoan parasites to cause more severe disease than the virus-free strains. The structure of LRV1 virus-like particles has been determined previously, however, the structure of the LRV1 virion has not been characterized. Here we used cryo-electron microscopy and single-particle reconstruction to determine the structures of the LRV1 virion and empty particle isolated from Leishmania guyanensis to resolutions of 4.0 Å and 3.6 Å, respectively. The capsid of LRV1 is built from sixty dimers of capsid proteins organized with icosahedral symmetry. RNA genomes of totiviruses are replicated inside the virions by RNA polymerases expressed as C-terminal extensions of a sub-population of capsid proteins. Most of the virions probably contain one or two copies of the RNA polymerase, however, the location of the polymerase domains in LRV1 capsid could not be identified, indicating that it varies among particles. Importance. Every year over 200 000 people contract leishmaniasis and more than five hundred people die of the disease. The mucocutaneous form of leishmaniasis produces lesions that can destroy the mucous membranes of the nose, mouth, and throat. Leishmania parasites carrying Leishmania RNA virus 1 (LRV1) are predisposed to cause aggravated symptoms in the mucocutaneous form of leishmaniasis. Here, we present the structure of the LRV1 virion determined using cryo-electron microscopy. #1:  ジャーナル: J Virol / 年: 2021 ジャーナル: J Virol / 年: 2021タイトル: Capsid Structure of Leishmania RNA virus 1 著者: Prochazkova M / Fuzik T / Grybchuk D / Falginella FL / Podesvova L / Yurchenko V / Vacha R / Plevka P | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_14566.map.gz emd_14566.map.gz | 59.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-14566-v30.xml emd-14566-v30.xml emd-14566.xml emd-14566.xml | 17.7 KB 17.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_14566_fsc.xml emd_14566_fsc.xml | 18.1 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_14566.png emd_14566.png | 169.6 KB | ||
| その他 |  emd_14566_half_map_1.map.gz emd_14566_half_map_1.map.gz emd_14566_half_map_2.map.gz emd_14566_half_map_2.map.gz | 408.5 MB 408.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14566 http://ftp.pdbj.org/pub/emdb/structures/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14566 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_14566_validation.pdf.gz emd_14566_validation.pdf.gz | 787.4 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_14566_full_validation.pdf.gz emd_14566_full_validation.pdf.gz | 787 KB | 表示 | |
| XML形式データ |  emd_14566_validation.xml.gz emd_14566_validation.xml.gz | 26.3 KB | 表示 | |
| CIF形式データ |  emd_14566_validation.cif.gz emd_14566_validation.cif.gz | 35.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14566 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7z90MC  7ns2C C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_14566.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_14566.map.gz / 形式: CCP4 / 大きさ: 512 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Virion structure | ||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.063 Å | ||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #2
| ファイル | emd_14566_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
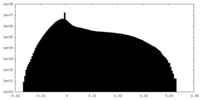
| 投影像・断面図 |
| ||||||||||||
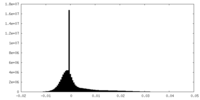
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_14566_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
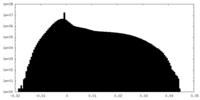
| 投影像・断面図 |
| ||||||||||||
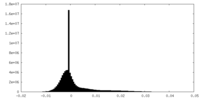
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Leishmania RNA virus 1 - 4
| 全体 | 名称:  Leishmania RNA virus 1 - 4 (ウイルス) Leishmania RNA virus 1 - 4 (ウイルス) |
|---|---|
| 要素 |
|
-超分子 #1: Leishmania RNA virus 1 - 4
| 超分子 | 名称: Leishmania RNA virus 1 - 4 / タイプ: virus / ID: 1 / 親要素: 0 / 含まれる分子: all / NCBI-ID: 12530 / 生物種: Leishmania RNA virus 1 - 4 / ウイルスタイプ: VIRION / ウイルス・単離状態: STRAIN / ウイルス・エンベロープ: No / ウイルス・中空状態: No |
|---|---|
| 宿主 | 生物種:  Leishmania guyanensis (ガイアナリーシュマニア) Leishmania guyanensis (ガイアナリーシュマニア)株: MHOM/BR/75/M4147 |
| 分子量 | 理論値: 9.72 MDa |
| ウイルス殻 | Shell ID: 1 / 名称: capsid / 直径: 420.0 Å / T番号(三角分割数): 2 |
-分子 #1: Capsid protein,Major capsid protein
| 分子 | 名称: Capsid protein,Major capsid protein / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Leishmania RNA virus 1 - 4 (ウイルス) Leishmania RNA virus 1 - 4 (ウイルス) |
| 分子量 | 理論値: 86.431742 KDa |
| 配列 | 文字列: MADIPNSDKI ACGRKPMFCE IIKLANRKRL IFNTTDERVY DARLNYCSTA DSTVQADCHI YWRLKLRRTD AVFEEYTGQG YSLDTAAYP QQYTDIIRGY YSKHVSSSLA ANTQHFVNVL AMLRHAGACI AHYCMTGKID FDILSKKKHK NKEVVTLSNA D SLSFLPHS ...文字列: MADIPNSDKI ACGRKPMFCE IIKLANRKRL IFNTTDERVY DARLNYCSTA DSTVQADCHI YWRLKLRRTD AVFEEYTGQG YSLDTAAYP QQYTDIIRGY YSKHVSSSLA ANTQHFVNVL AMLRHAGACI AHYCMTGKID FDILSKKKHK NKEVVTLSNA D SLSFLPHS ALYLPSPLRA SDPEIFNMLY LLGCACDASI AMDNISNTSG AAKYSMPHYN PLQLSHALHV TIFYMLSLMD SC GYGDDAV LALTSGLHSV TTVIAHSDEG GITRDALREL SYTQPYGTMP VPIAGYFQHI NVLFTTQPAW DQFAGIWDYV ILA TAALVH LSDPGMTVND VTYPTTLTTK VATVDGRNSD LAAQMMHSAT RFCDIFVENL STFWGVVANP DGNASQALLH AFNI VACAV EPNRHLEMNV MAPWYWVESS ALFCDYAPFR SPISSAGYGP QCVYGARLVL AATNSLEFTG EAGDYSAYRF EWTTM RHNP LFNILNKRVG DGLANVDFRL RPFNEWLLEG QPSRRSCNSA GHGTPTATCS HKTPNHDTLD EYIWGSTSCD LFHPAE LTS YTAVCVRFRN YLSGADGDVR ILNTPTREVI EGNVVTRCDG IRCLDSNKRI QHVPEVARRY CMMARYLAQA RTFGALT IG DDIIRGFDKV EKIVKMHKSN NRLDQMPLID VTGLCQPMIE TSTVRASTTT RIDPNKLAAA TARVELPLAP RCTSSLIP S SDTVPEPEPQ VGEPGDNGGC APDLGTGGGS GIEGRGSMDI GDPNSVQALA RLQASSVDKL AAALE |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 |
|---|---|
| 凍結 | 凍結剤: ETHANE-PROPANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON III (4k x 4k) 検出モード: COUNTING / 撮影したグリッド数: 1 / 実像数: 4542 / 平均電子線量: 1.98 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.2 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー




 Z
Z Y
Y X
X