[English] 日本語
 Yorodumi
Yorodumi- EMDB-13595: Helical reconstruction of TOROID (TORC1 Organized in Inhibited Do... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of TOROID (TORC1 Organized in Inhibited Domains) filaments. | |||||||||||||||
 Map data Map data | Helical reconstruction of TOROID (TORC1 Organized in Inhibited Domains) filaments. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 9.1 Å | |||||||||||||||
 Authors Authors | Felix J / Prouteau M / Bourgoint C / Bonnadei L / Desfosses A / Gabus C / Sadian Y / Savvides SN / Gutsche I / Loewith R | |||||||||||||||
| Funding support |  Switzerland, Switzerland,  France, 4 items France, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: EGOC inhibits TOROID polymerization by structurally activating TORC1. Authors: Manoël Prouteau / Clélia Bourgoint / Jan Felix / Lenny Bonadei / Yashar Sadian / Caroline Gabus / Savvas N Savvides / Irina Gutsche / Ambroise Desfosses / Robbie Loewith /    Abstract: Target of rapamycin complex 1 (TORC1) is a protein kinase controlling cell homeostasis and growth in response to nutrients and stresses. In Saccharomyces cerevisiae, glucose depletion triggers a ...Target of rapamycin complex 1 (TORC1) is a protein kinase controlling cell homeostasis and growth in response to nutrients and stresses. In Saccharomyces cerevisiae, glucose depletion triggers a redistribution of TORC1 from a dispersed localization over the vacuole surface into a large, inactive condensate called TOROID (TORC1 organized in inhibited domains). However, the mechanisms governing this transition have been unclear. Here, we show that acute depletion and repletion of EGO complex (EGOC) activity is sufficient to control TOROID distribution, independently of other nutrient-signaling pathways. The 3.9-Å-resolution structure of TORC1 from TOROID cryo-EM data together with interrogation of key interactions in vivo provide structural insights into TORC1-TORC1' and TORC1-EGOC interaction interfaces. These data support a model in which glucose-dependent activation of EGOC triggers binding to TORC1 at an interface required for TOROID assembly, preventing TORC1 polymerization and promoting release of active TORC1. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13595.map.gz emd_13595.map.gz | 22.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13595-v30.xml emd-13595-v30.xml emd-13595.xml emd-13595.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13595_fsc.xml emd_13595_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13595.png emd_13595.png | 3.8 MB | ||
| Others |  emd_13595_half_map_1.map.gz emd_13595_half_map_1.map.gz emd_13595_half_map_2.map.gz emd_13595_half_map_2.map.gz | 79.2 MB 79.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13595 http://ftp.pdbj.org/pub/emdb/structures/EMD-13595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13595 | HTTPS FTP |
-Validation report
| Summary document |  emd_13595_validation.pdf.gz emd_13595_validation.pdf.gz | 739.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13595_full_validation.pdf.gz emd_13595_full_validation.pdf.gz | 739.1 KB | Display | |
| Data in XML |  emd_13595_validation.xml.gz emd_13595_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_13595_validation.cif.gz emd_13595_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13595 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13595 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13595.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13595.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical reconstruction of TOROID (TORC1 Organized in Inhibited Domains) filaments. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||
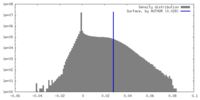
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Helical reconstruction of TOROID (TORC1 Organized in Inhibited...
| File | emd_13595_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical reconstruction of TOROID (TORC1 Organized in Inhibited Domains) filaments, half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
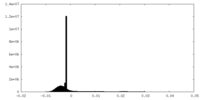
| Density Histograms |
-Half map: Helical reconstruction of TOROID (TORC1 Organized in Inhibited...
| File | emd_13595_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical reconstruction of TOROID (TORC1 Organized in Inhibited Domains) filaments, half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
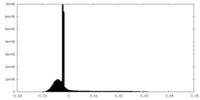
| Density Histograms |
- Sample components
Sample components
-Entire : TORC1 Organized in Inhibited Domains (TOROID)
| Entire | Name: TORC1 Organized in Inhibited Domains (TOROID) |
|---|---|
| Components |
|
-Supramolecule #1: TORC1 Organized in Inhibited Domains (TOROID)
| Supramolecule | Name: TORC1 Organized in Inhibited Domains (TOROID) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Target of rapamycin complex 1 subunit KOG1
| Macromolecule | Name: Target of rapamycin complex 1 subunit KOG1 / type: protein_or_peptide / ID: 1 Details: S. cerevisiae TOROIDs (TORC1 Organized in Inhibited Domains) in the sample are composed of Kog1 (P38873), Lst8 (P41318) and Tor2 (P32600). Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MPEIYGPQPL KPLNTVMRHG FEEQYQSDQL LQSLANDFIF YFDDKRHKTN GNPIPEEDKQ RDVNRYYQP ITDWKIMKDR QKTVSAALLL CLNLGVDPPD VMKTHPCARV EAWVDPLNFQ D SKKAIEQI GKNLQAQYET LSLRTRYKQS LDPCVEDVKR FCNSLRRTSK ...String: MPEIYGPQPL KPLNTVMRHG FEEQYQSDQL LQSLANDFIF YFDDKRHKTN GNPIPEEDKQ RDVNRYYQP ITDWKIMKDR QKTVSAALLL CLNLGVDPPD VMKTHPCARV EAWVDPLNFQ D SKKAIEQI GKNLQAQYET LSLRTRYKQS LDPCVEDVKR FCNSLRRTSK EDRILFHYNG HG VPKPTKS GEIWVFNRGY TQYIPVSLYD LQTWLGAPCI FVYDCNSAEN ILINFQKFVQ KRI KDDEEG NHDVAAPSPT SAYQDCFQLA SCTSDELLLM SPELPADLFS CCLTCPIEIS IRIF LMQSP LKDSKYKIFF ENSTSNQPFG DSKNSFKSKI PNVNIPGMLS DRRTPLGELN WIFTA ITDT IAWTSLPRPL FKKLFRHDLM IAALFRNFLL AKRIMPWYNC HPVSDPELPD SITTHP MWK SWDLAMDEVL TKIVIDLKNA PPATALESQM ILQQQETLQN GGSSKSNAQD TKAGSIQ TQ SRFAVANLST MSLVNNPALQ SRKSISLQSS QQQLQQQQQQ QQQFTGFFEQ NLTAFELW L KYASNVRHPP EQLPIVLQVL LSQVHRIRAL VLLSRFLDLG PWAVYLSLSI GIFPYVLKL LQSPAPELKP ILVFIWARIM SIDYKNTQSE LIKEKGYMYF VTVLVPDWGV NGMSATNGSA MINSGNPLT MTASQNINGP SSRYYERQQG NRTSNLGHNN LPFYHSNDTT DEQKAMAVFV L ASFVRNFP LGQKNCFSLE LVNKLCFYID NSEIPLLRQW CVILLGLLFA DNPLNRFVCM NT GAVEILL KSLKDPVPEV RTASIFALKH FISGFQDAEV ILRLQQEFEE QYQQLHSQLQ HLQ NQSHLQ QQQSQQQQQH LEQQQMKIEK QIRHCQVMQN QLEVIDLRKL KRQEIGNLIS ILPL INDGS SLVRKELVVY FSHIVSRYSN FFIVVVFNDL LEEIKLLEKS DINTRNTSDK YSVSQ GSIF YTVWKSLLIL AEDPFLENKE LSKQVIDYIL LELSAHKELG GPFAVMEKFL LKRSSK AHQ TGKFGFNSSQ VQFVKSSLRS FSPNERVDNN AFKKEQQQHD PKISHPMRTS LAKLFQS LG FSESNSDSDT QSSNTSMKSH TSKKGPSGLY LLNGNNNIYP TAETPRFRKH TEPLQLPL N SSFLDYSREY FQEPQMKKQE ADEPGSVEYN ARLWRRNRNE TIIQETQGEK KLSIYGNWS KKLISLNNKS QPKLMKFAQF EDQLITADDR STITVFDWEK GKTLSKFSNG TPFGTKVTDL KLINEDDSA LLLTGSSDGV IKIYRDYQDV DTFKIVSAWR GLTDMLLTPR STGLLTEWLQ I RGSLLTTG DVKVIRVWDA HTETVEVDIP AKTSSLITSL TADQLAGNIF VAGFADGSLR VY DRRLDPR DSMIRRWRAG NDKQGVWINN VHLQRGGYRE LVSGATNGVV ELWDIRSEDP VES FVDQNV TSQYGSQQKP TTMTCMQVHE HAPIIATGTK QIKIWTTSGD LLNSFKNSHN NGVT STLAA TGIPKSLSYS STSDAFLSSM AFHPHRMMIA ATNSHDSIVN IYKCEDERID YF |
-Macromolecule #2: Serine/threonine-protein kinase TOR2
| Macromolecule | Name: Serine/threonine-protein kinase TOR2 / type: protein_or_peptide / ID: 2 Details: S. cerevisiae TOROIDs (TORC1 Organized in Inhibited Domains) in the sample are composed of Kog1 (P38873), Lst8 (P41318) and Tor2 (P32600). Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MNKYINKYTT PPNLLSLRQR AEGKHRTRKK LTHKSHSHDD EMSTTSNTDS NHNGPNDSGR VITGSAGHI GKISFVDSEL DTTFSTLNLI FDKLKSDVPQ ERASGANELS TTLTSLAREV S AEQFQRFS NSLNNKIFEL IHGFTSSEKI GGILAVDTLI SFYLSTEELP ...String: MNKYINKYTT PPNLLSLRQR AEGKHRTRKK LTHKSHSHDD EMSTTSNTDS NHNGPNDSGR VITGSAGHI GKISFVDSEL DTTFSTLNLI FDKLKSDVPQ ERASGANELS TTLTSLAREV S AEQFQRFS NSLNNKIFEL IHGFTSSEKI GGILAVDTLI SFYLSTEELP NQTSRLANYL RV LIPSSDI EVMRLAANTL GRLTVPGGTL TSDFVEFEVR TCIDWLTLTA DNNSSSSKLE YRR HAALLI IKALADNSPY LLYPYVNSIL DNIWVPLRDA KLIIRLDAAV ALGKCLTIIQ DRDP ALGKQ WFQRLFQGCT HGLSLNTNDS VHATLLVFRE LLSLKAPYLR DKYDDIYKST MKYKE YKFD VIRREVYAIL PLLAAFDPAI FTKKYLDRIM VHYLRYLKNI DMNAANNSDK PFILVS IGD IAFEVGSSIS PYMTLILDNI REGLRTKFKV RKQFEKDLFY CIGKLACALG PAFAKHL NK DLLNLMLNCP MSDHMQETLM ILNEKIPSLE STVNSRILNL LSISLSGEKF IQSNQYDF N NQFSIEKARK SRNQSFMKKT GESNDDITDA QILIQCFKML QLIHHQYSLT EFVRLITIS YIEHEDSSVR KLAALTSCDL FIKDDICKQT SVHALHSVSE VLSKLLMIAI TDPVAEIRLE ILQHLGSNF DPQLAQPDNL RLLFMALNDE IFGIQLEAIK IIGRLSSVNP AYVVPSLRKT L LELLTQLK FSNMPKKKEE SATLLCTLIN SSDEVAKPYI DPILDVILPK CQDASSAVAS TA LKVLGEL SVVGGKEMTR YLKELMPLII NTFQDQSNSF KRDAALTTLG QLAASSGYVV GPL LDYPEL LGILINILKT ENNPHIRRGT VRLIGILGAL DPYKHREIEV TSNSKSSVEQ NAPS IDIAL LMQGVSPSND EYYPTVVIHN LMKILNDPSL SIHHTAAIQA IMHIFQNLGL RCVSF LDQI IPGIILVMRS CPPSQLDFYF QQLGSLISIV KQHIRPHVEK IYGVIREFFP IIKLQI TII SVIESISKAL EGEFKRFVPE TLTFFLDILE NDQSNKRIVP IRILKSLVTF GPNLEDY SH LIMPIVVRMT EYSAGSLKKI SIITLGRLAK NINLSEMSSR IVQALVRILN NGDRELTK A TMNTLSLLLL QLGTDFVVFV PVINKALLRN RIQHSVYDQL VNKLLNNECL PTNIIFDKE NEVPERKNYE DEMQVTKLPV NQNILKNAWY CSQQKTKEDW QEWIRRLSIQ LLKESPSACL RSCSSLVSV YYPLARELFN ASFSSCWVEL QTSYQEDLIQ ALCKALSSSE NPPEIYQMLL N LVEFMEHD DKPLPIPIHT LGKYAQKCHA FAKALHYKEV EFLEEPKNST IEALISINNQ LH QTDSAIG ILKHAQQHNE LQLKETWYEK LQRWEDALAA YNEKEAAGED SVEVMMGKLR SLY ALGEWE ELSKLASEKW GTAKPEVKKA MAPLAAGAAW GLEQWDEIAQ YTSVMKSQSP DKEF YDAIL CLHRNNFKKA EVHIFNARDL LVTELSALVN ESYNRAYNVV VRAQIIAELE EIIKY KKLP QNSDKRLTMR ETWNTRLLGC QKNIDVWQRI LRVRSLVIKP KEDAQVRIKF ANLCRK SGR MALAKKVLNT LLEETDDPDH PNTAKASPPV VYAQLKYLWA TGLQDEALKQ LINFTSR MA HDLGLDPNNM IAQSVPQQSK RVPRHVEDYT KLLARCFLKQ GEWRVCLQPK WRLSNPDS I LGSYLLATHF DNTWYKAWHN WALANFEVIS MLTSVSKKKQ EGSDASSVTD INEFDNGMI GVNTFDAKEV HYSSNLIHRH VIPAIKGFFH SISLSESSSL QDALRLLTLW FTFGGIPEAT QAMHEGFNL IQIGTWLEVL PQLISRIHQP NQIVSRSLLS LLSDLGKAHP QALVYPLMVA I KSESLSRQ KAALSIIEKM RIHSPVLVDQ AELVSHELIR MAVLWHEQWY EGLDDASRQF FG EHNTEKM FAALEPLYEM LKRGPETLRE ISFQNSFGRD LNDAYEWLMN YKKSKDVSNL NQA WDIYYN VFRKIGKQLP QLQTLELQHV SPKLLSAHDL ELAVPGTRAS GGKPIVKISK FEPV FSVIS SKQRPRKFCI KGSDGKDYKY VLKGHEDIRQ DSLVMQLFGL VNTLLQNDAE CFRRH LDIQ QYPAIPLSPK SGLLGWVPNS DTFHVLIREH REAKKIPLNI EHWVMLQMAP DYDNLT LLQ KVEVFTYALN NTEGQDLYKV LWLKSRSSET WLERRTTYTR SLAVMSMTGY ILGLGDR HP SNLMLDRITG KVIHIDFGDC FEAAILREKF PEKVPFRLTR MLTYAMEVSG IEGSFRIT C ENVMKVLRDN KGSLMAILEA FAFDPLINWG FDLPTKKIEE ETGIQLPVMN ANELLSNGA ITEEEVQRVE NEHKNAIRNA RAMLVLKRIT DKLTGNDIRR FNDLDVPEQV DKLIQQATSV ENLCQHYIG WCPFW |
-Macromolecule #3: Target of rapamycin complex subunit LST8
| Macromolecule | Name: Target of rapamycin complex subunit LST8 / type: protein_or_peptide / ID: 3 Details: S. cerevisiae TOROIDs (TORC1 Organized in Inhibited Domains) in the sample are composed of Kog1 (P38873), Lst8 (P41318) and Tor2 (P32600). Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSVILVSAGY DHTIRFWEAL TGVCSRTIQH SDSQVNRLEI TNDKKLLATA GHQNVRLYDI RTTNPNPVA SFEGHRGNVT SVSFQQDNRW MVTSSEDGTI KVWDVRSPSI PRNYKHNAPV N EVVIHPNQ GELISCDRDG NIRIWDLGEN QCTHQLTPED DTSLQSLSMA ...String: MSVILVSAGY DHTIRFWEAL TGVCSRTIQH SDSQVNRLEI TNDKKLLATA GHQNVRLYDI RTTNPNPVA SFEGHRGNVT SVSFQQDNRW MVTSSEDGTI KVWDVRSPSI PRNYKHNAPV N EVVIHPNQ GELISCDRDG NIRIWDLGEN QCTHQLTPED DTSLQSLSMA SDGSMLAAAN TK GNCYVWE MPNHTDASHL KPVTKFRAHS TYITRILLSS DVKHLATCSA DHTARVWSID DDF KLETTL DGHQRWVWDC AFSADSAYLV TASSDHYVRL WDLSTREIVR QYGGHHKGAV CVAL NDV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 50 mM MES-NaOH pH 6.0, 5 mM CHAPS, 600 mM KCl, 0,5 mM DTT, Phosphatase Inhibitor Mix, Complete Protease Inhibitor Cocktail (Roche; 1 tablet/ 50 ml) and 1 mM PMSF) | ||||||||||||||||||||||||
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-16 / Number real images: 4901 / Average exposure time: 8.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 37000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)