[English] 日本語
 Yorodumi
Yorodumi- EMDB-13485: Cryo-EM structure of Bestrhodopsin (rhodopsin-rhodopsin-bestrophi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Bestrhodopsin (rhodopsin-rhodopsin-bestrophin) complex | |||||||||
 Map data Map data | Sharpened cryoEM map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Phaeocystis antarctica (eukaryote) / Phaeocystis antarctica (eukaryote) /  Phaeocystis (eukaryote) Phaeocystis (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Matzov D / Kaczmarczyk I / Shalev-Benami M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Rhodopsin-bestrophin fusion proteins from unicellular algae form gigantic pentameric ion channels. Authors: Andrey Rozenberg / Igor Kaczmarczyk / Donna Matzov / Johannes Vierock / Takashi Nagata / Masahiro Sugiura / Kota Katayama / Yuma Kawasaki / Masae Konno / Yujiro Nagasaka / Mako Aoyama / ...Authors: Andrey Rozenberg / Igor Kaczmarczyk / Donna Matzov / Johannes Vierock / Takashi Nagata / Masahiro Sugiura / Kota Katayama / Yuma Kawasaki / Masae Konno / Yujiro Nagasaka / Mako Aoyama / Ishita Das / Efrat Pahima / Jonathan Church / Suliman Adam / Veniamin A Borin / Ariel Chazan / Sandra Augustin / Jonas Wietek / Julien Dine / Yoav Peleg / Akira Kawanabe / Yuichiro Fujiwara / Ofer Yizhar / Mordechai Sheves / Igor Schapiro / Yuji Furutani / Hideki Kandori / Keiichi Inoue / Peter Hegemann / Oded Béjà / Moran Shalev-Benami /    Abstract: Many organisms sense light using rhodopsins, photoreceptive proteins containing a retinal chromophore. Here we report the discovery, structure and biophysical characterization of bestrhodopsins, a ...Many organisms sense light using rhodopsins, photoreceptive proteins containing a retinal chromophore. Here we report the discovery, structure and biophysical characterization of bestrhodopsins, a microbial rhodopsin subfamily from marine unicellular algae, in which one rhodopsin domain of eight transmembrane helices or, more often, two such domains in tandem, are C-terminally fused to a bestrophin channel. Cryo-EM analysis of a rhodopsin-rhodopsin-bestrophin fusion revealed that it forms a pentameric megacomplex (~700 kDa) with five rhodopsin pseudodimers surrounding the channel in the center. Bestrhodopsins are metastable and undergo photoconversion between red- and green-absorbing or green- and UVA-absorbing forms in the different variants. The retinal chromophore, in a unique binding pocket, photoisomerizes from all-trans to 11-cis form. Heterologously expressed bestrhodopsin behaves as a light-modulated anion channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13485.map.gz emd_13485.map.gz | 223.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13485-v30.xml emd-13485-v30.xml emd-13485.xml emd-13485.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13485.png emd_13485.png | 143.5 KB | ||
| Others |  emd_13485_additional_1.map.gz emd_13485_additional_1.map.gz emd_13485_half_map_1.map.gz emd_13485_half_map_1.map.gz emd_13485_half_map_2.map.gz emd_13485_half_map_2.map.gz | 191.1 MB 194.4 MB 194.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13485 http://ftp.pdbj.org/pub/emdb/structures/EMD-13485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13485 | HTTPS FTP |
-Validation report
| Summary document |  emd_13485_validation.pdf.gz emd_13485_validation.pdf.gz | 808.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13485_full_validation.pdf.gz emd_13485_full_validation.pdf.gz | 808.1 KB | Display | |
| Data in XML |  emd_13485_validation.xml.gz emd_13485_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_13485_validation.cif.gz emd_13485_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13485 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13485 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13485 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13485 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13485.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13485.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened cryoEM map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
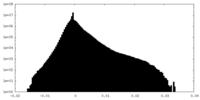
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened cryoEM map
| File | emd_13485_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryoEM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
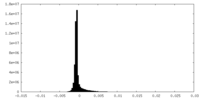
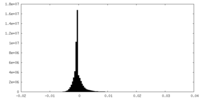
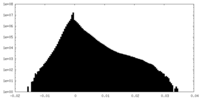
| Density Histograms |
-Half map: half map 1
| File | emd_13485_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
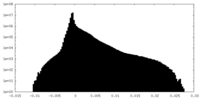
| Density Histograms |
-Half map: half map 2
| File | emd_13485_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
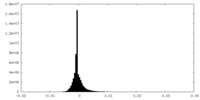
| Density Histograms |
- Sample components
Sample components
-Entire : Rhodopsin
| Entire | Name: Rhodopsin |
|---|---|
| Components |
|
-Supramolecule #1: Rhodopsin
| Supramolecule | Name: Rhodopsin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Phaeocystis antarctica (eukaryote) Phaeocystis antarctica (eukaryote) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 670 KDa |
-Macromolecule #1: Rhodopsin
| Macromolecule | Name: Rhodopsin / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Phaeocystis (eukaryote) Phaeocystis (eukaryote) |
| Molecular weight | Theoretical: 134.01025 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASSTAAPPA AVPAIETAAP PSPEPRDSVS VNEGGDASSA QAGAAVGSTI IVGAPELEKE YSDLNFAQVA RDEGRRCLLM CVAFAIAIA HLYIYPALFG VRIVDQAEVP AEERTYFHHG WTAMLVIFFI EGVTVFLKVC STRKTRWLEK AVLQKLDGNI G VLIGEYIV ...String: MASSTAAPPA AVPAIETAAP PSPEPRDSVS VNEGGDASSA QAGAAVGSTI IVGAPELEKE YSDLNFAQVA RDEGRRCLLM CVAFAIAIA HLYIYPALFG VRIVDQAEVP AEERTYFHHG WTAMLVIFFI EGVTVFLKVC STRKTRWLEK AVLQKLDGNI G VLIGEYIV VAATYIIMGA NLIPVFEEPR SGRRVYAVRY MEWTIDACGL VYLDCRILFG MPFSKFRMLL VYSVLYMLFG LW AALASTW MWYAIFLSAS WFFFGLVCYY YWTFHRQNPS PLQQFGRAPI KQAILVFVIV WWVLYGVLFM LCFQAPDVVP QWL EQLLWT GMDVVMKLSH TVVLMAWRET QWEIDAVVDR QKVEAGRAIA QLDHQRAIHE RDLVRLRSRV YYGEHIKSEE EIKS EEVIS RSLRARKSRQ GQDGTEPSSA LSGASSTKEP PALEENSGSA APSWTAVLAK GKATSSPFAR VNKIFMREAG LCLVL CLAF VVALLHLPVY SEWFGVEVLD AEAVPHDELG FFHHGWTTML VVFLIESITV LLKVWSTWHD PRLAENVAQQ LSGNLG VLI AEYLVVGATY VILGYNLMPV FVVHRPGVAS RRVYAVRYME WAVDATGLIW LDCHCLFSRN FNEFRMAIVW TVAYMLF GL WSALASTWAW YWAFLLASWA AFLIVCLILV RFLRQDPYPH QPFGKTSVKP CILAFIIGWW VLYGILFMVC FQAPDAVP Q WLEQFLWTGM DVVMKLSHTV VLMAWRTTEW NVCELHGRNQ AEKSKKSLLN EGSHARQEKQ QLEAIMLEGG GVEAAGRVG SVSFAPSSSR RRKESDSTNW TATPGLRVDL SSMVRLEGQL AQGLVTDVHR KGMMRSEDLA ELKRLEESGF LQAQQHRNWE SQTREMTFL AHGINHIAYD PRSWMKTLTA VRGRAPTSFL LWVVLIESSI VLALSKFFGE SFDLGVSSGI HSLFGVLVSF L VVFRTQAA FKKWWSGRSA VSSLVQMSRT FAQQVCAYVK DEAYVNRMVR YSIATVVATR CHLRNTRIDP AMLLGVLKEE EI EELNRQK NLPFYTAWVI RSTLAEAVAE GACLPLHMAI ENAIKAIEQS IADAERLLTP MPFTYVVHVR TFLFIYLMGL PFI LVEDLG WLMLVAVSFL GYLMIGLENT AVQLENPFGT DCNHHPLDLY CLEVSQDLLH LLDLRASAKA Q |
-Macromolecule #2: RETINAL
| Macromolecule | Name: RETINAL / type: ligand / ID: 2 / Number of copies: 5 / Formula: RET |
|---|---|
| Molecular weight | Theoretical: 284.436 Da |
| Chemical component information |  ChemComp-RET: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 0.86 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.21 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 27749 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X