+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of E. coli MutL bound to a 3' resected DNA end | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA mismatch repair / Protein-DNA complex / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsingle-stranded DNA-dependent ATP-dependent DNA helicase complex / mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication / mismatch repair complex / regulation of DNA recombination / : / mismatched DNA binding / ATP-dependent DNA damage sensor activity / mismatch repair / ATP hydrolysis activity / DNA binding ...single-stranded DNA-dependent ATP-dependent DNA helicase complex / mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication / mismatch repair complex / regulation of DNA recombination / : / mismatched DNA binding / ATP-dependent DNA damage sensor activity / mismatch repair / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species | DNA molecule (others) /   | |||||||||
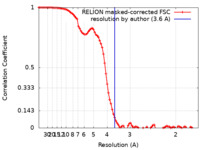
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Borsellini A / Lamers MH | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: MutL binds to 3' resected DNA ends and blocks DNA polymerase access. Authors: Alessandro Borsellini / Joyce H G Lebbink / Meindert H Lamers /  Abstract: DNA mismatch repair removes mis-incorporated bases after DNA replication and reduces the error rate a 100-1000-fold. After recognition of a mismatch, a large section of up to a thousand nucleotides ...DNA mismatch repair removes mis-incorporated bases after DNA replication and reduces the error rate a 100-1000-fold. After recognition of a mismatch, a large section of up to a thousand nucleotides is removed from the daughter strand followed by re-synthesis. How these opposite activities are coordinated is poorly understood. Here we show that the Escherichia coli MutL protein binds to the 3' end of the resected strand and blocks access of Pol I and Pol III. The cryo-EM structure of an 85-kDa MutL-DNA complex, determined to 3.7 Å resolution, reveals a unique DNA binding mode that positions MutL at the 3' end of a primer-template, but not at a 5' resected DNA end or a blunt DNA end. Hence, our work reveals a novel role for MutL in the final stages of mismatch repair by preventing premature DNA synthesis during removal of the mismatched strand. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13255.map.gz emd_13255.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13255-v30.xml emd-13255-v30.xml emd-13255.xml emd-13255.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13255_fsc.xml emd_13255_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_13255.png emd_13255.png | 123.6 KB | ||
| Filedesc metadata |  emd-13255.cif.gz emd-13255.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13255 http://ftp.pdbj.org/pub/emdb/structures/EMD-13255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13255 | HTTPS FTP |
-Validation report
| Summary document |  emd_13255_validation.pdf.gz emd_13255_validation.pdf.gz | 369.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13255_full_validation.pdf.gz emd_13255_full_validation.pdf.gz | 369.4 KB | Display | |
| Data in XML |  emd_13255_validation.xml.gz emd_13255_validation.xml.gz | 10.7 KB | Display | |
| Data in CIF |  emd_13255_validation.cif.gz emd_13255_validation.cif.gz | 14 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13255 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13255 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13255 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13255 | HTTPS FTP |
-Related structure data
| Related structure data |  7p8vMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13255.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13255.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.859 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : E. coli MutL N-terminal dimer bound to a 3' resected DNA end
| Entire | Name: E. coli MutL N-terminal dimer bound to a 3' resected DNA end |
|---|---|
| Components |
|
-Supramolecule #1: E. coli MutL N-terminal dimer bound to a 3' resected DNA end
| Supramolecule | Name: E. coli MutL N-terminal dimer bound to a 3' resected DNA end type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: 3' resected DNA end
| Supramolecule | Name: 3' resected DNA end / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
-Supramolecule #3: E. coli MutL N-terminal dimer
| Supramolecule | Name: E. coli MutL N-terminal dimer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA mismatch repair protein MutL
| Macromolecule | Name: DNA mismatch repair protein MutL / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 68.005508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPIQVLPPQL ANQIAAGEVV ERPASVVKEL VENSLDAGAT RIDIDIERGG AKLIRIRDNG CGIKKDELAL ALARHATSKI ASLDDLEAI ISLGFRGEAL ASISSVSRLT LTSRTAEQQE AWQAYAEGRD MNVTVKPAAH PVGTTLEVLD LFYNTPARRK F LRTEKTEF ...String: MPIQVLPPQL ANQIAAGEVV ERPASVVKEL VENSLDAGAT RIDIDIERGG AKLIRIRDNG CGIKKDELAL ALARHATSKI ASLDDLEAI ISLGFRGEAL ASISSVSRLT LTSRTAEQQE AWQAYAEGRD MNVTVKPAAH PVGTTLEVLD LFYNTPARRK F LRTEKTEF NHIDEIIRRI ALARFDVTIN LSHNGKIVRQ YRAVPEGGQK ERRLGAICGT AFLEQALAIE WQHGDLTLRG WV ADPNHTT PALAEIQYCY VNGRMMRDRL INHAIRQACE DKLGADQQPA FVLYLEIDPH QVDVNVHPAK HEVRFHQSRL VHD FIYQGV LSVLQQQLET PLPLDDEPQP APRSIPENRV AAGRNHFAEP AAREPVAPRY TPAPASGSRP AAPWPNAQPG YQKQ QGEVY RQLLQTPAPM QKLKAPEPQE PALAANSQSF GRVLTIVHSD CALLERDGNI SLLSLPVAER WLRQAQLTPG EAPVC AQPL LIPLRLKVSA EEKSALEKAQ SALAELGIDF QSDAQHVTIR AVPLPLRQQN LQILIPELIG YLAKQSVFEP GNIAQW IAR NLMSEHAQWS MAQAITLLAD VERLCPQLVK TPPGGLLQSV DLHPAIKALK DE UniProtKB: DNA mismatch repair protein MutL |
-Macromolecule #2: Template strand
| Macromolecule | Name: Template strand / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 6.841428 KDa |
| Sequence | String: (DG)(DC)(DT)(DG)(DG)(DA)(DG)(DG)(DC)(DT) (DA)(DA)(DG)(DC)(DT)(DA)(DA)(DG)(DC)(DT) (DG)(DA) |
-Macromolecule #3: Primer strand
| Macromolecule | Name: Primer strand / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 3.941584 KDa |
| Sequence | String: (DT)(DC)(DA)(DG)(DC)(DT)(DT)(DA)(DG)(DC) (DT)(DT)(DA) |
-Macromolecule #4: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.0002 kPa | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 76 % / Chamber temperature: 277.15 K / Instrument: LEICA PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 539000 / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)