[English] 日本語
 Yorodumi
Yorodumi- EMDB-13244: PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S rib... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13244 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state III | |||||||||
 Map data Map data | PoxtA bound to 70S ribosome from E. faecalis, state III. From RELION Refine3d. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / Enterococcus faecalis / PoxtA / ABCF / antibiotic resistance protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / large ribosomal subunit / regulation of translation / ribosomal small subunit assembly / large ribosomal subunit rRNA binding / small ribosomal subunit / 5S rRNA binding / transferase activity / cytosolic small ribosomal subunit ...ribosomal small subunit biogenesis / small ribosomal subunit rRNA binding / large ribosomal subunit / regulation of translation / ribosomal small subunit assembly / large ribosomal subunit rRNA binding / small ribosomal subunit / 5S rRNA binding / transferase activity / cytosolic small ribosomal subunit / ribosomal large subunit assembly / cytosolic large ribosomal subunit / tRNA binding / rRNA binding / ribosome / structural constituent of ribosome / translation / ribonucleoprotein complex / mRNA binding / zinc ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
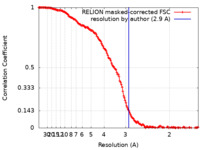
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Crowe-McAuliffe C / Wilson DN | |||||||||
| Funding support |  Germany, Germany,  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation | Journal: mSphere / Year: 2018 Title: The Enterococcus Cassette Chromosome, a Genomic Variation Enabler in Enterococci. Authors: A Sivertsen / J Janice / T Pedersen / T M Wagner / J Hegstad / K Hegstad /  Abstract: has a highly variable genome prone to recombination and horizontal gene transfer. Here, we have identified a novel genetic island with an insertion locus and mobilization genes similar to those of ... has a highly variable genome prone to recombination and horizontal gene transfer. Here, we have identified a novel genetic island with an insertion locus and mobilization genes similar to those of staphylococcus cassette chromosome elements SCC This novel element termed the enterococcus cassette chromosome (ECC) element was located in the 3' region of and encoded large serine recombinases similar to SCC Horizontal transfer of an ECC element termed ECC:: containing a knock-in chloramphenicol resistance determinant occurred in the presence of a conjugative plasmid. We determined the ECC:: insertion site in the 3' region of in the recipient by long-read sequencing. ECC:: also mobilized by homologous recombination through sequence identity between flanking insertion sequence (IS) elements in ECC:: and the conjugative plasmid. The genes were found in 69 of 516 genomes in GenBank. Full-length ECC elements were retrieved from 32 of these genomes. ECCs were flanked by and sites of approximately 50 bp. The sequences were found by PCR and sequencing of circularized ECCs in three strains. The genes in ECCs contained an amalgam of common and rare genes. Taken together, our data imply that ECC elements act as hot spots for genetic exchange and contribute to the large variation of accessory genes found in is a bacterium found in a great variety of environments, ranging from the clinic as a nosocomial pathogen to natural habitats such as mammalian intestines, water, and soil. They are known to exchange genetic material through horizontal gene transfer and recombination, leading to great variability of accessory genes and aiding environmental adaptation. Identifying mobile genetic elements causing sequence variation is important to understand how genetic content variation occurs. Here, a novel genetic island, the enterococcus cassette chromosome, is shown to contain a wealth of genes, which may aid in adapting to new environments. The transmission mechanism involves the only two conserved genes within ECC, , large serine recombinases that insert ECC into the host genome similarly to SCC elements found in staphylococci. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Structural basis for PoxtA-mediated resistance to Phenicol and Oxazolidinone antibiotics Authors: Crowe-McAuliffe C / Murina V / Kasari M / Takada H / Turnbull KJ / Sundsfjord A / Hegstad K / Atkinson GC / Wilson DN / Hauryliuk V | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13244.map.gz emd_13244.map.gz | 252 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13244-v30.xml emd-13244-v30.xml emd-13244.xml emd-13244.xml | 87.6 KB 87.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13244_fsc.xml emd_13244_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_13244.png emd_13244.png | 126 KB | ||
| Masks |  emd_13244_msk_1.map emd_13244_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13244.cif.gz emd-13244.cif.gz | 15.8 KB | ||
| Others |  emd_13244_additional_1.map.gz emd_13244_additional_1.map.gz emd_13244_half_map_1.map.gz emd_13244_half_map_1.map.gz emd_13244_half_map_2.map.gz emd_13244_half_map_2.map.gz | 42.5 MB 253.3 MB 253.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13244 http://ftp.pdbj.org/pub/emdb/structures/EMD-13244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13244 | HTTPS FTP |
-Validation report
| Summary document |  emd_13244_validation.pdf.gz emd_13244_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13244_full_validation.pdf.gz emd_13244_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_13244_validation.xml.gz emd_13244_validation.xml.gz | 22.6 KB | Display | |
| Data in CIF |  emd_13244_validation.cif.gz emd_13244_validation.cif.gz | 29.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13244 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13244 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13244 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13244 | HTTPS FTP |
-Related structure data
| Related structure data |  7p7tMC  7p7qC  7p7rC  7p7sC  7p7uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13244.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13244.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PoxtA bound to 70S ribosome from E. faecalis, state III. From RELION Refine3d. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
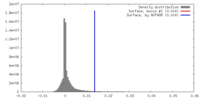
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13244_msk_1.map emd_13244_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
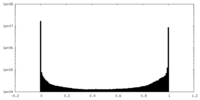
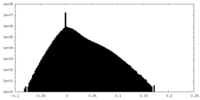
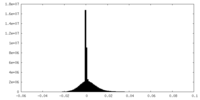
| Density Histograms |
-Additional map: PoxtA bound to 70S ribosome from E. faecalis,...
| File | emd_13244_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PoxtA bound to 70S ribosome from E. faecalis, state III. Sharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
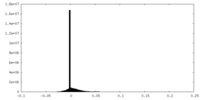
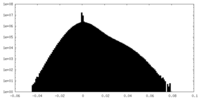
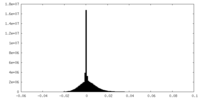
| Density Histograms |
-Half map: PoxtA bound to 70S ribosome from E. faecalis, state III. Half map.
| File | emd_13244_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PoxtA bound to 70S ribosome from E. faecalis, state III. Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: PoxtA bound to 70S ribosome from E. faecalis, state III. Half map.
| File | emd_13244_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PoxtA bound to 70S ribosome from E. faecalis, state III. Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of PoxtA-EQ2 bound to the 70S ribosome from E. faecalis (...
+Supramolecule #1: Complex of PoxtA-EQ2 bound to the 70S ribosome from E. faecalis (...
+Supramolecule #2: ARE-ABC-F family resistance factor PoxtA
+Supramolecule #4: A-site tRNA
+Supramolecule #3: 70S Ribosome
+Macromolecule #1: ARE-ABC-F family resistance factor PoxtA
+Macromolecule #2: 50S ribosomal protein L29
+Macromolecule #3: 50S ribosomal protein L30
+Macromolecule #4: 50S ribosomal protein L31 type B
+Macromolecule #5: 50S ribosomal protein L32
+Macromolecule #6: 50S ribosomal protein L33
+Macromolecule #7: 50S ribosomal protein L34
+Macromolecule #8: 50S ribosomal protein L35
+Macromolecule #9: 50S ribosomal protein L36
+Macromolecule #14: 50S ribosomal protein L1
+Macromolecule #15: 50S ribosomal protein L2
+Macromolecule #16: 50S ribosomal protein L3
+Macromolecule #17: 50S ribosomal protein L4
+Macromolecule #18: 50S ribosomal protein L5
+Macromolecule #19: 50S ribosomal protein L6
+Macromolecule #20: 50S ribosomal protein L13
+Macromolecule #21: 50S ribosomal protein L14
+Macromolecule #22: 50S ribosomal protein L15
+Macromolecule #23: 50S ribosomal protein L16
+Macromolecule #24: 50S ribosomal protein L17
+Macromolecule #25: 50S ribosomal protein L18
+Macromolecule #26: 50S ribosomal protein L19
+Macromolecule #27: 50S ribosomal protein L20
+Macromolecule #28: 50S ribosomal protein L21
+Macromolecule #29: 50S ribosomal protein L22
+Macromolecule #30: 50S ribosomal protein L23
+Macromolecule #31: 50S ribosomal protein L24
+Macromolecule #32: 50S ribosomal protein L27
+Macromolecule #33: 50S ribosomal protein L28
+Macromolecule #36: 30S ribosomal protein S2
+Macromolecule #37: 30S ribosomal protein S3
+Macromolecule #38: 30S ribosomal protein S4
+Macromolecule #39: 30S ribosomal protein S5
+Macromolecule #40: 30S ribosomal protein S6
+Macromolecule #41: 30S ribosomal protein S7
+Macromolecule #42: 30S ribosomal protein S8
+Macromolecule #43: 30S ribosomal protein S9
+Macromolecule #44: 30S ribosomal protein S10
+Macromolecule #45: 30S ribosomal protein S11
+Macromolecule #46: 30S ribosomal protein S12
+Macromolecule #47: 30S ribosomal protein S13
+Macromolecule #48: 30S ribosomal protein S14 type Z
+Macromolecule #49: 30S ribosomal protein S15
+Macromolecule #50: 30S ribosomal protein S16
+Macromolecule #51: 30S ribosomal protein S17
+Macromolecule #52: 30S ribosomal protein S18
+Macromolecule #53: 30S ribosomal protein S19
+Macromolecule #54: 30S ribosomal protein S20
+Macromolecule #10: 23S rRNA
+Macromolecule #11: 5S rRNA
+Macromolecule #12: A-site tRNA
+Macromolecule #13: tRNA-fMet
+Macromolecule #34: 16S rRNA
+Macromolecule #35: mRNA
+Macromolecule #55: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #56: MAGNESIUM ION
+Macromolecule #57: ZINC ION
+Macromolecule #58: POTASSIUM ION
+Macromolecule #59: SPECTINOMYCIN
+Macromolecule #60: 1,4-DIAMINOBUTANE
+Macromolecule #61: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 9 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK II |
| Details | Grid was prepared by four applications of elution fraction from affinity purification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 3639 / Average exposure time: 5.0 sec. / Average electron dose: 30.255 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)