[English] 日本語
 Yorodumi
Yorodumi- EMDB-1231: Structure of the connector of bacteriophage T7 at 8A resolution: ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1231 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
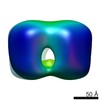
| Title | Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. | |||||||||
 Map data Map data | 3D map of T7-connector | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 8.0 Å | |||||||||
 Authors Authors | Agirrezabala X / Martin-Benito J / Valle M / Gonzalez JM / Valencia A / Valpuesta JM / Carrascosa JL | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2005 Journal: J Mol Biol / Year: 2005Title: Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. Authors: Xabier Agirrezabala / Jaime Martín-Benito / Mikel Valle / José M González / Alfonso Valencia / José María Valpuesta / José L Carrascosa /  Abstract: The three-dimensional structure of the bacteriophage T7 head-to-tail connector has been obtained at 8A resolution using cryo-electron microscopy and single-particle analysis from purified recombinant ...The three-dimensional structure of the bacteriophage T7 head-to-tail connector has been obtained at 8A resolution using cryo-electron microscopy and single-particle analysis from purified recombinant connectors. The general morphology of the T7 connector is that of a 12-folded toroidal homopolymer with a channel that runs along the longitudinal axis of the particle. The structure of the T7 connector reveals many structural similarities with the connectors from other bacteriophages. Docking of the atomic structure of the varphi29 connector into the three-dimensional reconstruction of T7 connector reveals that the narrow, distal region of the two oligomers are almost identical. This region of the varphi29 connector has been suggested to be involved in DNA translocation, and is composed of an alpha-beta-alpha-beta-beta-alpha motif. A search for alpha-helices in the same region of the T7 three-dimensional map has located three alpha-helices in approximately the same position as those of the varphi29 connector. A comparison of the predicted secondary structure of several bacteriophage connectors, including among others T7, varphi29, P22 and SPP1, reveals that, despite the lack of sequence homology, they seem to contain the same alpha-beta-alpha-beta-beta-alpha motif as that present in the varphi29 connector. These results allow us to suggest a common architecture related to a basic component of the DNA translocating machinery for several viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1231.map.gz emd_1231.map.gz | 9.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1231-v30.xml emd-1231-v30.xml emd-1231.xml emd-1231.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1231.gif 1231.gif | 38.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1231 http://ftp.pdbj.org/pub/emdb/structures/EMD-1231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1231 | HTTPS FTP |
-Validation report
| Summary document |  emd_1231_validation.pdf.gz emd_1231_validation.pdf.gz | 218.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1231_full_validation.pdf.gz emd_1231_full_validation.pdf.gz | 217.7 KB | Display | |
| Data in XML |  emd_1231_validation.xml.gz emd_1231_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1231 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1231 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1231 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1231 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1231.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1231.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of T7-connector | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.18 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Connector of Bacteriophage T7
+Supramolecule #1000: Connector of Bacteriophage T7
+Macromolecule #1: gp8
+Macromolecule #2: gp8
+Macromolecule #3: gp8
+Macromolecule #4: gp8
+Macromolecule #5: gp8
+Macromolecule #6: gp8
+Macromolecule #7: gp8
+Macromolecule #8: gp8
+Macromolecule #9: gp8
+Macromolecule #10: gp8
+Macromolecule #11: gp8
+Macromolecule #12: gp8
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.8 Details: 20mM Tris HCl pH:7.8, 3mM B-mercaptoethanol, 4%(v/v) glicerol, 2mM Na3EDTA, 100 mM NaCl |
| Staining | Type: NEGATIVE / Details: cryoEM |
| Grid | Details: Quantifoil grids with 2 um hole size |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: FFT with ssCCD |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Average electron dose: 10 e/Å2 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 64220 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 2.3 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder, GATAN. Eucentric Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Wiener filter, defocus groups |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C12 (12 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 26492 |
| Final angle assignment | Details: SPIDER |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















