+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12186 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of KdpFABC in E2Pi state with BeF3 and K+ | ||||||||||||
 Map data Map data | non-uniform refinement by cryoSPARC | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationP-type K+ transporter / potassium:proton antiporter complex / potassium ion-transporting ATPase complex / P-type potassium transmembrane transporter activity / monoatomic cation transmembrane transport /  potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding / potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding /  ATP hydrolysis activity ...P-type K+ transporter / potassium:proton antiporter complex / potassium ion-transporting ATPase complex / P-type potassium transmembrane transporter activity / monoatomic cation transmembrane transport / ATP hydrolysis activity ...P-type K+ transporter / potassium:proton antiporter complex / potassium ion-transporting ATPase complex / P-type potassium transmembrane transporter activity / monoatomic cation transmembrane transport /  potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding / potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / ATP binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||
| Biological species |   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) | ||||||||||||
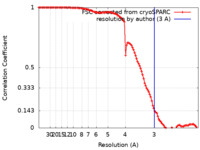
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Sweet ME / Larsen C / Pedersen BP / Stokes DL | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Structural basis for potassium transport in prokaryotes by KdpFABC. Authors: Marie E Sweet / Casper Larsen / Xihui Zhang / Michael Schlame / Bjørn P Pedersen / David L Stokes /   Abstract: KdpFABC is an oligomeric K transport complex in prokaryotes that maintains ionic homeostasis under stress conditions. The complex comprises a channel-like subunit (KdpA) from the superfamily of K ...KdpFABC is an oligomeric K transport complex in prokaryotes that maintains ionic homeostasis under stress conditions. The complex comprises a channel-like subunit (KdpA) from the superfamily of K transporters and a pump-like subunit (KdpB) from the superfamily of P-type ATPases. Recent structural work has defined the architecture and generated contradictory hypotheses for the transport mechanism. Here, we use substrate analogs to stabilize four key intermediates in the reaction cycle and determine the corresponding structures by cryogenic electron microscopy. We find that KdpB undergoes conformational changes consistent with other representatives from the P-type superfamily, whereas KdpA, KdpC, and KdpF remain static. We observe a series of spherical densities that we assign as K or water and which define a pathway for K transport. This pathway runs through an intramembrane tunnel in KdpA and delivers ions to sites in the membrane domain of KdpB. Our structures suggest a mechanism where ATP hydrolysis is coupled to K transfer between alternative sites in KdpB, ultimately reaching a low-affinity site where a water-filled pathway allows release of K to the cytoplasm. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12186.map.gz emd_12186.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12186-v30.xml emd-12186-v30.xml emd-12186.xml emd-12186.xml | 28.4 KB 28.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12186_fsc.xml emd_12186_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12186.png emd_12186.png | 78.4 KB | ||
| Masks |  emd_12186_msk_1.map emd_12186_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_12186_half_map_1.map.gz emd_12186_half_map_1.map.gz emd_12186_half_map_2.map.gz emd_12186_half_map_2.map.gz | 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12186 http://ftp.pdbj.org/pub/emdb/structures/EMD-12186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12186 | HTTPS FTP |
-Related structure data
| Related structure data |  7bh2MC  7bgyC  7bh1C  7lc3C  7lc6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12186.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12186.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-uniform refinement by cryoSPARC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.079 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12186_msk_1.map emd_12186_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A from non-uniform refinement in cryoSPARC
| File | emd_12186_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A from non-uniform refinement in cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B from non-uniform refinement in cryoSPARC
| File | emd_12186_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B from non-uniform refinement in cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : KdpFABC in E2Pi state with BeF3 and K+
+Supramolecule #1: KdpFABC in E2Pi state with BeF3 and K+
+Macromolecule #1: Potassium-transporting ATPase potassium-binding subunit
+Macromolecule #2: Potassium-transporting ATPase ATP-binding subunit
+Macromolecule #3: Potassium-transporting ATPase KdpC subunit
+Macromolecule #4: Potassium-transporting ATPase KdpF subunit
+Macromolecule #5: POTASSIUM ION
+Macromolecule #6: (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)...
+Macromolecule #7: MAGNESIUM ION
+Macromolecule #8: BERYLLIUM TRIFLUORIDE ION
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: The beryllium fluoride in the buffer was added to the protein mixture containing all other buffer components in the form of a pre-incubated mixture, at a 1:10 volume, to produce the final ...Details: The beryllium fluoride in the buffer was added to the protein mixture containing all other buffer components in the form of a pre-incubated mixture, at a 1:10 volume, to produce the final concentrations 2.5 mM BeSO4 and 10 mM NaF. | ||||||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 50.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL of protein mixture applied to grid. Blot time 4 seconds, blot force 0, no wait before plunging.. | ||||||||||||||||||||||||
| Details | DM-solublized KdpFABC complex purified through SEC the same day as grid freezing. Peak fraction diluted to 4.5 mg/mL and complexed with magnesium fluoride solution for >30 min at room temperature prior to application to grid. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Digitization - Sampling interval: 5.0 µm / Number grids imaged: 1 / Number real images: 2567 / Average exposure time: 2.5 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z
Z Y
Y X
X