[English] 日本語
 Yorodumi
Yorodumi- EMDB-11869: Structure of the actin filament Arp2/3 complex branch junction in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11869 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the actin filament Arp2/3 complex branch junction in cells | |||||||||
 Map data Map data | Structure of the actin filament Arp2/3 complex branch junction | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Actin / Arp2-3 complex / Cytoskeleton / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpodosome core / concave side of sperm head / actin filament branch point / ventral surface of cell / microtubule organizing center localization / negative regulation of bleb assembly / positive regulation of barbed-end actin filament capping / apical tubulobulbar complex / tubulobulbar complex / regulation of myosin II filament organization ...podosome core / concave side of sperm head / actin filament branch point / ventral surface of cell / microtubule organizing center localization / negative regulation of bleb assembly / positive regulation of barbed-end actin filament capping / apical tubulobulbar complex / tubulobulbar complex / regulation of myosin II filament organization / meiotic chromosome movement towards spindle pole / cytosolic transport / growth cone leading edge / peripheral region of growth cone / muscle cell projection membrane / : / meiotic cytokinesis / apical ectoplasmic specialization / basal ectoplasmic specialization / cellular response to rapamycin / lamellipodium organization / spindle localization / : / skeletal muscle fiber adaptation / leading edge of lamellipodium / Striated Muscle Contraction / EPHB-mediated forward signaling / cellular response to trichostatin A / RHO GTPases Activate WASPs and WAVEs / Arp2/3 protein complex / hemidesmosome / asymmetric cell division / protein kinase C signaling / Arp2/3 complex-mediated actin nucleation / actin filament network formation / podosome ring / orbitofrontal cortex development / actin cap / postsynaptic actin cytoskeleton organization / Regulation of actin dynamics for phagocytic cup formation / negative regulation of axon extension / maintenance of cell polarity / Clathrin-mediated endocytosis / positive regulation of dendritic spine morphogenesis / positive regulation of astrocyte differentiation / : / apical dendrite / astrocyte differentiation / positive regulation of fibroblast migration / podosome / positive regulation of synapse assembly / positive regulation of podosome assembly / positive regulation of dendrite morphogenesis / response to steroid hormone / positive regulation of filopodium assembly / smooth muscle cell migration / establishment or maintenance of cell polarity / positive regulation of actin filament polymerization / cellular response to platelet-derived growth factor stimulus / mesenchyme migration / positive regulation of smooth muscle cell migration / cell leading edge / filamentous actin / brush border / associative learning / skeletal muscle thin filament assembly / striated muscle thin filament / excitatory synapse / response to immobilization stress / positive regulation of double-strand break repair via homologous recombination / cilium assembly / positive regulation of protein targeting to membrane / glutamate receptor binding / skeletal muscle fiber development / positive regulation of lamellipodium assembly / stress fiber / response to mechanical stimulus / positive regulation of substrate adhesion-dependent cell spreading / axon terminus / cellular response to epidermal growth factor stimulus / cellular response to transforming growth factor beta stimulus / ruffle / cytoskeletal protein binding / positive regulation of neuron differentiation / actin filament polymerization / Neutrophil degranulation / cellular response to nerve growth factor stimulus / secretory granule / meiotic cell cycle / filopodium / dendritic shaft / cell projection / actin filament / positive regulation of protein localization to plasma membrane / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / structural constituent of cytoskeleton / cellular response to type II interferon / ruffle membrane / actin filament binding / cell-cell junction Similarity search - Function | |||||||||
| Biological species |  | |||||||||
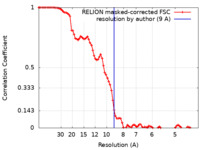
| Method | subtomogram averaging / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Faessler F / Dimchev G | |||||||||
| Funding support |  Austria, 1 items Austria, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Authors: Florian Fäßler / Georgi Dimchev / Victor-Valentin Hodirnau / William Wan / Florian K M Schur /   Abstract: The actin-related protein (Arp)2/3 complex nucleates branched actin filament networks pivotal for cell migration, endocytosis and pathogen infection. Its activation is tightly regulated and involves ...The actin-related protein (Arp)2/3 complex nucleates branched actin filament networks pivotal for cell migration, endocytosis and pathogen infection. Its activation is tightly regulated and involves complex structural rearrangements and actin filament binding, which are yet to be understood. Here, we report a 9.0 Å resolution structure of the actin filament Arp2/3 complex branch junction in cells using cryo-electron tomography and subtomogram averaging. This allows us to generate an accurate model of the active Arp2/3 complex in the branch junction and its interaction with actin filaments. Notably, our model reveals a previously undescribed set of interactions of the Arp2/3 complex with the mother filament, significantly different to the previous branch junction model. Our structure also indicates a central role for the ArpC3 subunit in stabilizing the active conformation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11869.map.gz emd_11869.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11869-v30.xml emd-11869-v30.xml emd-11869.xml emd-11869.xml | 36.7 KB 36.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11869_fsc.xml emd_11869_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11869.png emd_11869.png | 39.9 KB | ||
| Masks |  emd_11869_msk_1.map emd_11869_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11869.cif.gz emd-11869.cif.gz | 8.4 KB | ||
| Others |  emd_11869_half_map_1.map.gz emd_11869_half_map_1.map.gz emd_11869_half_map_2.map.gz emd_11869_half_map_2.map.gz | 27 MB 27 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11869 http://ftp.pdbj.org/pub/emdb/structures/EMD-11869 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11869 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11869 | HTTPS FTP |
-Related structure data
| Related structure data |  7aqkMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11869.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11869.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the actin filament Arp2/3 complex branch junction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.137 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
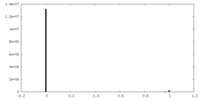
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11869_msk_1.map emd_11869_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
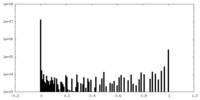
| Density Histograms |
-Half map: Half map 1
| File | emd_11869_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
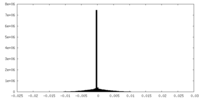
| Density Histograms |
-Half map: Half map 2
| File | emd_11869_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
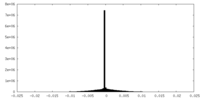
| Density Histograms |
- Sample components
Sample components
-Entire : Actin Filament Arp2/3 Complex Branch Junction
| Entire | Name: Actin Filament Arp2/3 Complex Branch Junction |
|---|---|
| Components |
|
-Supramolecule #1: Actin Filament Arp2/3 Complex Branch Junction
| Supramolecule | Name: Actin Filament Arp2/3 Complex Branch Junction / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all Details: Structure obtained from the actin network of extracted and fixed mouse fibroblast lamellipodia |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin-related protein 2, Arp2
| Macromolecule | Name: Actin-related protein 2, Arp2 / type: protein_or_peptide / ID: 1 Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they ...Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.818711 KDa |
| Sequence | String: MDSQGRKVVV CDNGTGFVKC GYAGSNFPEH IFPALVGRPI IRSTTKVGNI EIKDLMVGDE ASELRSMLEV NYPMENGIVR NWDDMKHLW DYTFGPEKLN IDTRNCKILL TEPPMNPTKN REKIVEVMFE TYQFSGVYVA IQAVLTLYAQ GLLTGVVVDS G DGVTHICP ...String: MDSQGRKVVV CDNGTGFVKC GYAGSNFPEH IFPALVGRPI IRSTTKVGNI EIKDLMVGDE ASELRSMLEV NYPMENGIVR NWDDMKHLW DYTFGPEKLN IDTRNCKILL TEPPMNPTKN REKIVEVMFE TYQFSGVYVA IQAVLTLYAQ GLLTGVVVDS G DGVTHICP VYEGFSLPHL TRRLDIAGRD ITRYLIKLLL LRGYAFNHSA DFETVRMIKE KLCYVGYNIE QEQKLALETT VL VESYTLP DGRIIKVGGE RFEAPEALFQ PHLINVEGVG VAELLFNTIQ AADIDTRSEF YKHIVLSGGS TMYPGLPSRL ERE LKQLYL ERVLKGDVEK LSKFKIRIED PPRRKHMVFL GGAVLADIMK DKDNFWMTRQ EYQEKGVRVL EKLGVTVR |
-Macromolecule #2: Actin-related protein 3, Arp3
| Macromolecule | Name: Actin-related protein 3, Arp3 / type: protein_or_peptide / ID: 2 Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they ...Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.428031 KDa |
| Sequence | String: MAGRLPACVV DCGTGYTKLG YAGNTEPQFI IPSCIAIKES AKVGDQAQRR VMKGVDDLDF FIGDEAIEKP TYATKWPIRH GIVEDWDLM ERFMEQVIFK YLRAEPEDHY FLLTEPPLNT PENREYTAEI MFESFNVPGL YIAVQAVLAL AASWTSRQVG E RTLTGTVI ...String: MAGRLPACVV DCGTGYTKLG YAGNTEPQFI IPSCIAIKES AKVGDQAQRR VMKGVDDLDF FIGDEAIEKP TYATKWPIRH GIVEDWDLM ERFMEQVIFK YLRAEPEDHY FLLTEPPLNT PENREYTAEI MFESFNVPGL YIAVQAVLAL AASWTSRQVG E RTLTGTVI DSGDGVTHVI PVAEGYVIGS CIKHIPIAGR DITYFIQQLL RDREVGIPPE QSLETAKAVK ERYSYVCPDL VK EFNKYDT DGSKWIKQYT GINAISKKEF SIDVGYERFL GPEIFFHPEF ANPDFTQPIS EVVDEVIQNC PIDVRRPLYK NIV LSGGST MFRDFGRRLQ RDLKRTVDAR LKLSEELSGG RLKPKPIDVQ VITHHMQRYA VWFGGSMLAS TPEFYQVCHT KKDY EEIGP SICRHNPVFG VMS |
-Macromolecule #3: Actin-related protein 2/3 complex subunit 1b, ArpC1b
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 1b, ArpC1b / type: protein_or_peptide / ID: 3 Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they ...Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.016738 KDa |
| Sequence | String: MAYHSFLVEP ISCHAWNKDR TQIAICPNNH EVHIYEKSGN KWVQVHELKE HNGQVTGVDW APDSNRIVTC GTDRNAYVWT LKGRTWKPT LVILRINRAA RCVRWAPNEK KFAVGSGSRV ISICYFEQEN DWWVCKHIKK PIRSTVLSLD WHPNSVLLAA G SCDFKCRI ...String: MAYHSFLVEP ISCHAWNKDR TQIAICPNNH EVHIYEKSGN KWVQVHELKE HNGQVTGVDW APDSNRIVTC GTDRNAYVWT LKGRTWKPT LVILRINRAA RCVRWAPNEK KFAVGSGSRV ISICYFEQEN DWWVCKHIKK PIRSTVLSLD WHPNSVLLAA G SCDFKCRI FSAYIKEVEE RPAPTPWGSK MPFGELMFES SSSCGWVHGV CFSANGSRVA WVSHDSTVCL ADADKKMAVA TL ASETLPL LAVTFITESS LVAAGHDCFP VLFTYDSAAG KLSFGGRLDV PKQSSQRGLT ARERFQNLDK KASSEGSAAA GAG LDSLHK NSVSQISVLS GGKAKCSQFC TTGMDGGMSI WDVRSLESAL KDLKIV |
-Macromolecule #4: Actin-related protein 2/3 complex subunit 2, ArpC2
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 2, ArpC2 / type: protein_or_peptide / ID: 4 Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they ...Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.402043 KDa |
| Sequence | String: MILLEVNNRI IEETLALKFE NAAAGNKPEA VEVTFADFDG VLYHISNPNG DKTKVMVSIS LKFYKELQAH GADELLKRVY GSYLVNPES GYNVSLLYDL ENLPASKDSI VHQAGMLKRN CFASVFEKYF QFQEEGKEGE NRAVIHYRDD ETMYVESKKD R VTVVFSTV ...String: MILLEVNNRI IEETLALKFE NAAAGNKPEA VEVTFADFDG VLYHISNPNG DKTKVMVSIS LKFYKELQAH GADELLKRVY GSYLVNPES GYNVSLLYDL ENLPASKDSI VHQAGMLKRN CFASVFEKYF QFQEEGKEGE NRAVIHYRDD ETMYVESKKD R VTVVFSTV FKDDDDVVIG KVFMQEFKEG RRASHTAPQV LFSHREPPLE LKDTDAAVGD NIGYITFVLF PRHTNASARD NT INLIHTF RDYLHYHIKC SKAYIHTRMR AKTSDFLKVL NRARPDAEKK EMKTITGKTF SSR |
-Macromolecule #5: Actin-related protein 2/3 complex subunit 3, ArpC3
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 3, ArpC3 / type: protein_or_peptide / ID: 5 Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they ...Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.572666 KDa |
| Sequence | String: MPAYHSSLMD PDTKLIGNMA LLPIRSQFKG PAPRETKDTD IVDEAIYYFK ANVFFKNYEI KNEADRTLIY ITLYISECLK KLQKCNSKS QGEKEMYTLG ITNFPIPGEP GFPLNAIYAK PANKQEDEVM RAYLQQLRQE TGLRLCEKVF DPQNDKPSKW W TCFVKRQF MNKSLSGPGQ |
-Macromolecule #6: Actin-related protein 2/3 complex subunit 4, ArpC4
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 4, ArpC4 / type: protein_or_peptide / ID: 6 Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they ...Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.697047 KDa |
| Sequence | String: MTATLRPYLS AVRATLQAAL CLENFSSQVV ERHNKPEVEV RSSKELLLQP VTISRNEKEK VLIEGSINSV RVSIAVKQAD EIEKILCHK FMRFMMMRAE NFFILRRKPV EGYDISFLIT NFHTEQMYKH KLVDFVIHFM EEIDKEISEM KLSVNARARI V AEEFLKNF |
-Macromolecule #7: Actin-related protein 2/3 complex subunit 5, ArpC5
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 5, ArpC5 / type: protein_or_peptide / ID: 7 Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they ...Details: As models derived from Bos taurus Arp2/3 complexes have been used for fitting, also the Bos taurus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.295317 KDa |
| Sequence | String: MSKNTVSSAR FRKVDVDEYD ENKFVDEDDG GDGQAGPDEG EVDSCLRQGN MTAALQAALK NPPINTKSQA VKDRAGSIVL KVLISFKAN DIEKAVQSLD KNGVDLLMKY IYKGFESPSD NSSAVLLQWH EKALAAGGVG SIVRVLTARK TV |
-Macromolecule #8: Actin, alpha skeletal muscle, ACTA1
| Macromolecule | Name: Actin, alpha skeletal muscle, ACTA1 / type: protein_or_peptide / ID: 8 Details: As models derived from Oryctolagus cuniculus actin have been used for fitting, also the Oryctolagus cuniculus sequence (as well as the corresponding UniProt identifier) is given here, even ...Details: As models derived from Oryctolagus cuniculus actin have been used for fitting, also the Oryctolagus cuniculus sequence (as well as the corresponding UniProt identifier) is given here, even though they were fit into a mouse structure. Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.096953 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSSSL EK SYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM YPGIADRMQK EIT ALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6.1 Component:
Details: Adjust to pH 6.1 using NaOH | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR Details: After glow discharging of the grid and prior to the seeding of cells, the grid was coated using 25ug/ml Fibronectin | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K Details: Leica GP2, 3,5sec back-blotting, sensor on, 0,1mm movement after contact, manually pre-blotted within the chamber prior to the application of fiducials. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 1.21 sec. / Average electron dose: 2.79 e/Å2 Details: Images were collected in movie-mode at 7 frames per tilt |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -0.0055 µm / Nominal defocus min: -0.00175 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||||||||||||||||||||||||||||||||||||
| Output model |  PDB-7aqk: |
 Movie
Movie Controller
Controller






















 Z
Z Y
Y X
X