[English] 日本語
 Yorodumi
Yorodumi- EMDB-1164: Maturation of phage T7 involves structural modification of both s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1164 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Maturation of phage T7 involves structural modification of both shell and inner core components. | |||||||||
 Map data Map data | bacteriophage T7 mature virion | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 21.0 Å | |||||||||
 Authors Authors | Agirrezabala X / Martin-Benito J / Caston JR / Miranda R / Valpuesta JM / Carrascosa JL | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2005 Journal: EMBO J / Year: 2005Title: Maturation of phage T7 involves structural modification of both shell and inner core components. Authors: Xabier Agirrezabala / Jaime Martín-Benito / José R Castón / Roberto Miranda / José María Valpuesta / José L Carrascosa /  Abstract: The double-stranded DNA bacteriophages are good model systems to understand basic biological processes such as the macromolecular interactions that take place during the virus assembly and ...The double-stranded DNA bacteriophages are good model systems to understand basic biological processes such as the macromolecular interactions that take place during the virus assembly and maturation, or the behavior of molecular motors that function during the DNA packaging process. Using cryoelectron microscopy and single-particle methodology, we have determined the structures of two phage T7 assemblies produced during its morphogenetic process, the DNA-free prohead and the mature virion. The first structure reveals a complex assembly in the interior of the capsid, which involves the scaffolding, and the core complex, which plays an important role in DNA packaging and is located in one of the phage vertices. The reconstruction of the mature virion reveals important changes in the shell, now much larger and thinner, the disappearance of the scaffolding structure, and important rearrangements of the core complex, which now protrudes the shell and interacts with the tail. Some of these changes must originate by the pressure exerted by the DNA in the interior of the head. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1164.map.gz emd_1164.map.gz | 37.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1164-v30.xml emd-1164-v30.xml emd-1164.xml emd-1164.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  1164.gif 1164.gif | 8.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1164 http://ftp.pdbj.org/pub/emdb/structures/EMD-1164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1164 | HTTPS FTP |
-Validation report
| Summary document |  emd_1164_validation.pdf.gz emd_1164_validation.pdf.gz | 262.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1164_full_validation.pdf.gz emd_1164_full_validation.pdf.gz | 261.9 KB | Display | |
| Data in XML |  emd_1164_validation.xml.gz emd_1164_validation.xml.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1164 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1164 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1164 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1164 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1164.map.gz / Format: CCP4 / Size: 39.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1164.map.gz / Format: CCP4 / Size: 39.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | bacteriophage T7 mature virion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.66 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
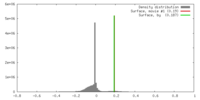
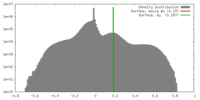
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : phage T7 mature virion
+Supramolecule #1000: phage T7 mature virion
+Macromolecule #1: gp14
+Macromolecule #2: gp15
+Macromolecule #3: gp16
+Macromolecule #4: gp8
+Macromolecule #5: gp10A
+Macromolecule #6: gp10B
+Macromolecule #7: gp11
+Macromolecule #8: gp12
+Macromolecule #9: gp17
+Macromolecule #10: genome
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.7 / Details: 50mM Tris-HCl pH:7.7 10mM MgCl2 100mM NaCl |
|---|---|
| Grid | Details: Quantifoil grids 2/2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: FFT with ssCCD |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Average electron dose: 10 e/Å2 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 29930 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder. GATAN. Eucentric Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Wiener filter, defocus groups |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Details: 5-fold symmetrya / Number images used: 4785 |
 Movie
Movie Controller
Controller


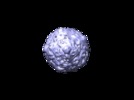








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)