[English] 日本語
 Yorodumi
Yorodumi- EMDB-1102: Three-dimensional structure and regulation of the DNA-dependent p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1102 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). | |||||||||
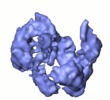
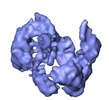
 Map data Map data | This is the cryoEM 3D reconstruction of DNA-PKcs kinase. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Rivera-Calzada A / Maman JP / Spagnolo L / Pearl LH / Llorca O | |||||||||
 Citation Citation |  Journal: Structure / Year: 2005 Journal: Structure / Year: 2005Title: Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Authors: Angel Rivera-Calzada / Joseph D Maman / Laura Spagnolo / Laurence H Pearl / Oscar Llorca /  Abstract: DNA-PKcs is a large PI3-kinase-related protein kinase (PIKK) that plays a central role in DNA double-strand break (DSB) repair via nonhomologous end joining. Using cryo-electron microscopy we have ...DNA-PKcs is a large PI3-kinase-related protein kinase (PIKK) that plays a central role in DNA double-strand break (DSB) repair via nonhomologous end joining. Using cryo-electron microscopy we have now generated an approximately 13 A three-dimensional map of DNA-PKcs, revealing the overall architecture and topology of the 4128 residue polypeptide chain and allowing location of domains. The highly conserved C-terminal PIKK catalytic domain forms a central structure from which FAT and FATC domains protrude. Conformational changes observed in these domains on DNA binding suggest that they transduce DNA-induced conformational changes to the catalytic core and regulate kinase activity. The N-terminal segments form long curved tubular-shaped domains based on helical repeats to create interacting surfaces required for macromolecular assembly. Comparison of DNA-PKcs with another PIKK DNA repair factor, ATM, defines a common architecture for this important protein family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1102.map.gz emd_1102.map.gz | 155 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1102-v30.xml emd-1102-v30.xml emd-1102.xml emd-1102.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1102.gif 1102.gif emd_1102.tif emd_1102.tif | 50.3 KB 263.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1102 http://ftp.pdbj.org/pub/emdb/structures/EMD-1102 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1102 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1102 | HTTPS FTP |
-Validation report
| Summary document |  emd_1102_validation.pdf.gz emd_1102_validation.pdf.gz | 199.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1102_full_validation.pdf.gz emd_1102_full_validation.pdf.gz | 199 KB | Display | |
| Data in XML |  emd_1102_validation.xml.gz emd_1102_validation.xml.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1102 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1102 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1102 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1102 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1102.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1102.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the cryoEM 3D reconstruction of DNA-PKcs kinase. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human DNA-dependent protein kinase catalytic subunit, DNA-PKcs
| Entire | Name: Human DNA-dependent protein kinase catalytic subunit, DNA-PKcs |
|---|---|
| Components |
|
-Supramolecule #1000: Human DNA-dependent protein kinase catalytic subunit, DNA-PKcs
| Supramolecule | Name: Human DNA-dependent protein kinase catalytic subunit, DNA-PKcs type: sample / ID: 1000 / Oligomeric state: monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 470 KDa / Method: From its sequence |
-Macromolecule #1: DNA-dependent protein kinase catalytic subunit
| Macromolecule | Name: DNA-dependent protein kinase catalytic subunit / type: protein_or_peptide / ID: 1 / Name.synonym: DNA-PKcs / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: HeLa / Organelle: Nucleus / Location in cell: Nucleus Homo sapiens (human) / synonym: Human / Cell: HeLa / Organelle: Nucleus / Location in cell: Nucleus |
| Molecular weight | Experimental: 470 KDa |
| Recombinant expression | Organism: HeLa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50mM Tris-HCl, 50mM KCl, 1mM MgCl2. |
| Grid | Details: 400 mesh carbon coated Rhodium-Copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 90 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: home made plunger / Method: Blot for 1 second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Temperature | Min: 90 K / Max: 90 K / Average: 90 K |
| Alignment procedure | Legacy - Astigmatism: objective astigmatism was corrected using the stigmator, CCD camera collected images and the Digital Micrograph software |
| Details | Microscope model: TECNAI G2 200 kV, FEI. |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 43 / Details: Images were averaged to 14 microns / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.3 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: side entry liquid-nitrogen cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: reverse phases for each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 13.0 Å / Resolution method: OTHER / Software - Name: EMAN Details: CTF Corrected particles were subjected to 3D refinement as implemented in EMAN. The starting volume was generated by image classification of the whole data set into a few average images to ...Details: CTF Corrected particles were subjected to 3D refinement as implemented in EMAN. The starting volume was generated by image classification of the whole data set into a few average images to build a reconstruction using common lines. Number images used: 7000 |
| Final angle assignment | Details: EMAN software criteria |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: SITUS |
| Details | Protocol: Rigid Body. The domains were fitted using the COLORES command from SITUS |
| Refinement | Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coeficient |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















