[English] 日本語
 Yorodumi
Yorodumi- EMDB-10935: Pinhead of the microtubule triplet of isolated Trichonympha agili... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10935 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pinhead of the microtubule triplet of isolated Trichonympha agilis centriole | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Trichonympha agilis (eukaryote) Trichonympha agilis (eukaryote) | |||||||||
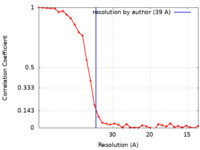
| Method | subtomogram averaging / cryo EM / Resolution: 39.0 Å | |||||||||
 Authors Authors | Nazarov S | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2020 Journal: EMBO J / Year: 2020Title: Novel features of centriole polarity and cartwheel stacking revealed by cryo-tomography. Authors: Sergey Nazarov / Alexandra Bezler / Georgios N Hatzopoulos / Veronika Nemčíková Villímová / Davide Demurtas / Maeva Le Guennec / Paul Guichard / Pierre Gönczy /  Abstract: Centrioles are polarized microtubule-based organelles that seed the formation of cilia, and which assemble from a cartwheel containing stacked ring oligomers of SAS-6 proteins. A cryo-tomography map ...Centrioles are polarized microtubule-based organelles that seed the formation of cilia, and which assemble from a cartwheel containing stacked ring oligomers of SAS-6 proteins. A cryo-tomography map of centrioles from the termite flagellate Trichonympha spp. was obtained previously, but higher resolution analysis is likely to reveal novel features. Using sub-tomogram averaging (STA) in T. spp. and Trichonympha agilis, we delineate the architecture of centriolar microtubules, pinhead, and A-C linker. Moreover, we report ~25 Å resolution maps of the central cartwheel, revealing notably polarized cartwheel inner densities (CID). Furthermore, STA of centrioles from the distant flagellate Teranympha mirabilis uncovers similar cartwheel architecture and a distinct filamentous CID. Fitting the CrSAS-6 crystal structure into the flagellate maps and analyzing cartwheels generated in vitro indicate that SAS-6 rings can directly stack onto one another in two alternating configurations: with a slight rotational offset and in register. Overall, improved STA maps in three flagellates enabled us to unravel novel architectural features, including of centriole polarity and cartwheel stacking, thus setting the stage for an accelerated elucidation of underlying assembly mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10935.map.gz emd_10935.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10935-v30.xml emd-10935-v30.xml emd-10935.xml emd-10935.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10935_fsc.xml emd_10935_fsc.xml | 3.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10935.png emd_10935.png | 162.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10935 http://ftp.pdbj.org/pub/emdb/structures/EMD-10935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10935 | HTTPS FTP |
-Validation report
| Summary document |  emd_10935_validation.pdf.gz emd_10935_validation.pdf.gz | 262.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10935_full_validation.pdf.gz emd_10935_full_validation.pdf.gz | 261.5 KB | Display | |
| Data in XML |  emd_10935_validation.xml.gz emd_10935_validation.xml.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10935 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10935 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10935 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10935 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10935.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10935.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.982 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Isolated basal body from Trichonympha agilis
| Entire | Name: Isolated basal body from Trichonympha agilis |
|---|---|
| Components |
|
-Supramolecule #1: Isolated basal body from Trichonympha agilis
| Supramolecule | Name: Isolated basal body from Trichonympha agilis / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Trichonympha agilis (eukaryote) Trichonympha agilis (eukaryote) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)