+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10589 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Filament structure of PfAct2 | |||||||||
 Map data Map data | Final sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated actin polymerization-dependent cell-to-cell migration in host / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / Neutrophil degranulation / microfilament motor activity / cytoskeleton organization / actin filament / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / actin cytoskeleton / hydrolase activity ...symbiont-mediated actin polymerization-dependent cell-to-cell migration in host / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / Neutrophil degranulation / microfilament motor activity / cytoskeleton organization / actin filament / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / actin cytoskeleton / hydrolase activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Vahokoski J / Calder LJ / Lopez AJ / Rosenthal PB / Kursula I | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The structure and biochemical properties of PfAct2 Authors: Lopez AJ / Vahokoski J / Tajedin L / Calder LJ / Rosenthal PB / Kursula I | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10589.map.gz emd_10589.map.gz | 6.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10589-v30.xml emd-10589-v30.xml emd-10589.xml emd-10589.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10589.png emd_10589.png | 96.7 KB | ||
| Others |  emd_10589_additional_1.map.gz emd_10589_additional_1.map.gz emd_10589_half_map_1.map.gz emd_10589_half_map_1.map.gz emd_10589_half_map_2.map.gz emd_10589_half_map_2.map.gz | 106.1 MB 106.1 MB 106.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10589 http://ftp.pdbj.org/pub/emdb/structures/EMD-10589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10589 | HTTPS FTP |
-Validation report
| Summary document |  emd_10589_validation.pdf.gz emd_10589_validation.pdf.gz | 371 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10589_full_validation.pdf.gz emd_10589_full_validation.pdf.gz | 370.2 KB | Display | |
| Data in XML |  emd_10589_validation.xml.gz emd_10589_validation.xml.gz | 12.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10589 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10589 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10589 | HTTPS FTP |
-Related structure data
| Related structure data |  8ccoM C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10589.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10589.map.gz / Format: CCP4 / Size: 134.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
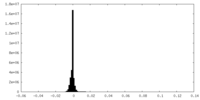
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: non-sharpened full map
| File | emd_10589_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-sharpened full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
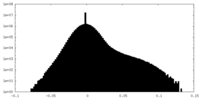
| Density Histograms |
-Half map: #1
| File | emd_10589_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
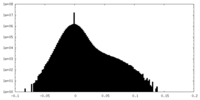
| Density Histograms |
-Half map: #2
| File | emd_10589_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Filamentous structure of PfAct2
| Entire | Name: Filamentous structure of PfAct2 |
|---|---|
| Components |
|
-Supramolecule #1: Filamentous structure of PfAct2
| Supramolecule | Name: Filamentous structure of PfAct2 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 47.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 28.37 Å Applied symmetry - Helical parameters - Δ&Phi: -166.931 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1 beta a7b0bd) / Number images used: 273310 |
|---|---|
| CTF correction | Software - Name: CTFFIND (ver. 4.1.10) |
| Startup model | Type of model: NONE / Details: a featureless cylinder |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)