+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10346 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of African Swine fever virus | ||||||||||||
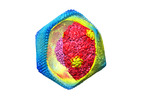
 Map data Map data | Postprocessed map | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  African swine fever virus BA71V African swine fever virus BA71V | ||||||||||||
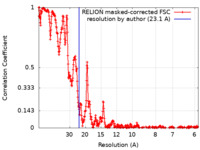
| Method | single particle reconstruction / cryo EM / Resolution: 23.1 Å | ||||||||||||
 Authors Authors | Abrescia NG / Andres G / Charro D / Matamoros T / Dillard R | ||||||||||||
| Funding support |  Spain, 3 items Spain, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2020 Journal: J Biol Chem / Year: 2020Title: The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. Authors: German Andrés / Diego Charro / Tania Matamoros / Rebecca S Dillard / Nicola G A Abrescia /   Abstract: African swine fever virus (ASFV) is a complex nucleocytoplasmic large DNA virus (NCLDV) that causes a devastating swine disease currently present in many countries of Africa, Europe, and Asia. ...African swine fever virus (ASFV) is a complex nucleocytoplasmic large DNA virus (NCLDV) that causes a devastating swine disease currently present in many countries of Africa, Europe, and Asia. Despite intense research efforts, relevant gaps in the architecture of the infectious virus particle remain. Here, we used single-particle cryo-EM to analyze the three-dimensional structure of the mature ASFV particle. Our results show that the ASFV virion, with a radial diameter of ∼2,080 Å, encloses a genome-containing nucleoid surrounded by two distinct icosahedral protein capsids and two lipoprotein membranes. The outer capsid forms a hexagonal lattice (triangulation number = 277) composed of 8,280 copies of the double jelly-roll major capsid protein (MCP) p72, arranged in trimers displaying a pseudo-hexameric morphology, and of 60 copies of a penton protein at the vertices. The inner protein layer, organized as a = 19 capsid, confines the core shell, and it is composed of the mature products derived from the ASFV polyproteins pp220 and pp62. Also, an icosahedral membrane lies between the two protein layers, whereas a pleomorphic envelope wraps the outer capsid. This high-level organization confers to ASFV a unique architecture among the NCLDVs that likely reflects the complexity of its infection process and may help explain current challenges in controlling it. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10346.map.gz emd_10346.map.gz | 3.4 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10346-v30.xml emd-10346-v30.xml emd-10346.xml emd-10346.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10346_fsc.xml emd_10346_fsc.xml | 35.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10346.png emd_10346.png | 515.1 KB | ||
| Masks |  emd_10346_msk_1.map emd_10346_msk_1.map | 3.7 GB |  Mask map Mask map | |
| Others |  emd_10346_additional_1.map.gz emd_10346_additional_1.map.gz emd_10346_additional_2.map.gz emd_10346_additional_2.map.gz emd_10346_half_map_1.map.gz emd_10346_half_map_1.map.gz emd_10346_half_map_2.map.gz emd_10346_half_map_2.map.gz | 504.4 MB 634.1 MB 3.4 GB 3.4 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10346 http://ftp.pdbj.org/pub/emdb/structures/EMD-10346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10346 | HTTPS FTP |
-Validation report
| Summary document |  emd_10346_validation.pdf.gz emd_10346_validation.pdf.gz | 410.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10346_full_validation.pdf.gz emd_10346_full_validation.pdf.gz | 409.5 KB | Display | |
| Data in XML |  emd_10346_validation.xml.gz emd_10346_validation.xml.gz | 35.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10346 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10346 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10346 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10346 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10346.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10346.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.938 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
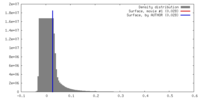
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10346_msk_1.map emd_10346_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
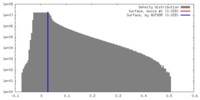
| Density Histograms |
-Additional map: nucleocapsid
| File | emd_10346_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nucleocapsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
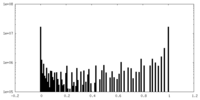
| Density Histograms |
-Additional map: simmetrized nucleocapsid
| File | emd_10346_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | simmetrized nucleocapsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
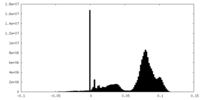
| Density Histograms |
-Half map: Half 2
| File | emd_10346_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half 1
| File | emd_10346_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : African swine fever virus BA71V
| Entire | Name:  African swine fever virus BA71V African swine fever virus BA71V |
|---|---|
| Components |
|
-Supramolecule #1: African swine fever virus BA71V
| Supramolecule | Name: African swine fever virus BA71V / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 10498 / Sci species name: African swine fever virus BA71V / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism: pigs, warthogs, bushpigs (unknown) |
| Virus shell | Shell ID: 1 / Name: Outer capsid (MCP p72) / Diameter: 2400.0 Å / T number (triangulation number): 277 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Formula: PBS |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Two datasets were acquired. |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: FEI FALCON III (4k x 4k) / #0 - Average electron dose: 47.7 e/Å2 #0 - Details: Titan Krios equipped with a Cs-corrector and a Falcon 3EC direct electron detector #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Average electron dose: 47.5 e/Å2 #1 - Details: Titan Krios equipped with a Gatan K2 Summit direct electron detector |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)