[English] 日本語
 Yorodumi
Yorodumi- EMDB-0176: Cryo-EM structure of a 70S Bacillus subtilis ribosome translating... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0176 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | |||||||||||||||
 Map data Map data | None | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | single particle cryo-EM / stalling peptide / ribosome / telithromycin | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of rRNA processing / nucleoid / rRNA processing / large ribosomal subunit / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit ...positive regulation of rRNA processing / nucleoid / rRNA processing / large ribosomal subunit / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / mRNA binding / DNA binding / RNA binding / zinc ion binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Crowe-McAuliffe C / Graf M | |||||||||||||||
| Funding support |  Germany, Germany,  Czech Republic, 4 items Czech Republic, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: Structural basis for antibiotic resistance mediated by the ABCF ATPase VmlR. Authors: Caillan Crowe-McAuliffe / Michael Graf / Paul Huter / Hiraku Takada / Maha Abdelshahid / Jiří Nováček / Victoriia Murina / Gemma C Atkinson / Vasili Hauryliuk / Daniel N Wilson /     Abstract: Many Gram-positive pathogenic bacteria employ ribosomal protection proteins (RPPs) to confer resistance to clinically important antibiotics. In , the RPP VmlR confers resistance to lincomycin (Lnc) ...Many Gram-positive pathogenic bacteria employ ribosomal protection proteins (RPPs) to confer resistance to clinically important antibiotics. In , the RPP VmlR confers resistance to lincomycin (Lnc) and the streptogramin A (S) antibiotic virginiamycin M (VgM). VmlR is an ATP-binding cassette (ABC) protein of the F type, which, like other antibiotic resistance (ARE) ABCF proteins, is thought to bind to antibiotic-stalled ribosomes and promote dissociation of the drug from its binding site. To investigate the molecular mechanism by which VmlR confers antibiotic resistance, we have determined a cryo-electron microscopy (cryo-EM) structure of an ATPase-deficient VmlR-EQ mutant in complex with a ErmDL-stalled ribosomal complex (SRC). The structure reveals that VmlR binds within the E site of the ribosome, with the antibiotic resistance domain (ARD) reaching into the peptidyltransferase center (PTC) of the ribosome and a C-terminal extension (CTE) making contact with the small subunit (SSU). To access the PTC, VmlR induces a conformational change in the P-site tRNA, shifting the acceptor arm out of the PTC and relocating the CCA end of the P-site tRNA toward the A site. Together with microbiological analyses, our study indicates that VmlR allosterically dissociates the drug from its ribosomal binding site and exhibits specificity to dislodge VgM, Lnc, and the pleuromutilin tiamulin (Tia), but not chloramphenicol (Cam), linezolid (Lnz), nor the macrolide erythromycin (Ery). | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0176.map.gz emd_0176.map.gz | 161.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0176-v30.xml emd-0176-v30.xml emd-0176.xml emd-0176.xml | 74.3 KB 74.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0176.png emd_0176.png | 104.9 KB | ||
| Filedesc metadata |  emd-0176.cif.gz emd-0176.cif.gz | 13.6 KB | ||
| Others |  emd_0176_additional.map.gz emd_0176_additional.map.gz | 106.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0176 http://ftp.pdbj.org/pub/emdb/structures/EMD-0176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0176 | HTTPS FTP |
-Validation report
| Summary document |  emd_0176_validation.pdf.gz emd_0176_validation.pdf.gz | 653.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0176_full_validation.pdf.gz emd_0176_full_validation.pdf.gz | 653.2 KB | Display | |
| Data in XML |  emd_0176_validation.xml.gz emd_0176_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_0176_validation.cif.gz emd_0176_validation.cif.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0176 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0176 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0176 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0176 | HTTPS FTP |
-Related structure data
| Related structure data |  6ha1MC  0177C  6ha8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0176.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0176.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: None
| File | emd_0176_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
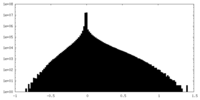
| Density Histograms |
- Sample components
Sample components
+Entire : 70S ribosome with P-tRNA translating the ErmD stalling peptide an...
+Supramolecule #1: 70S ribosome with P-tRNA translating the ErmD stalling peptide an...
+Supramolecule #2: 70S ribosome
+Supramolecule #3: ErmD stalling peptide
+Supramolecule #4: tRNA
+Macromolecule #1: 23S ribosomal RNA
+Macromolecule #2: 5S ribosomal RNA
+Macromolecule #30: mRNA
+Macromolecule #31: 16S ribosomal RNA
+Macromolecule #51: P-tRNA
+Macromolecule #3: 50S ribosomal protein L2
+Macromolecule #4: 50S ribosomal protein L3
+Macromolecule #5: 50S ribosomal protein L4
+Macromolecule #6: 50S ribosomal protein L5
+Macromolecule #7: 50S ribosomal protein L6
+Macromolecule #8: 50S ribosomal protein L13
+Macromolecule #9: 50S ribosomal protein L14
+Macromolecule #10: 50S ribosomal protein L15
+Macromolecule #11: 50S ribosomal protein L16
+Macromolecule #12: 50S ribosomal protein L17
+Macromolecule #13: 50S ribosomal protein L18
+Macromolecule #14: 50S ribosomal protein L19
+Macromolecule #15: 50S ribosomal protein L20
+Macromolecule #16: 50S ribosomal protein L21
+Macromolecule #17: 50S ribosomal protein L22
+Macromolecule #18: 50S ribosomal protein L23
+Macromolecule #19: 50S ribosomal protein L24
+Macromolecule #20: 50S ribosomal protein L27
+Macromolecule #21: 50S ribosomal protein L28
+Macromolecule #22: 50S ribosomal protein L29
+Macromolecule #23: 50S ribosomal protein L30
+Macromolecule #24: 50S ribosomal protein L32
+Macromolecule #25: 50S ribosomal protein L33 1
+Macromolecule #26: 50S ribosomal protein L34
+Macromolecule #27: 50S ribosomal protein L35
+Macromolecule #28: 50S ribosomal protein L36
+Macromolecule #29: 50S ribosomal protein L31
+Macromolecule #32: 30S ribosomal protein S2
+Macromolecule #33: 30S ribosomal protein S3
+Macromolecule #34: 30S ribosomal protein S4
+Macromolecule #35: 30S ribosomal protein S5
+Macromolecule #36: 30S ribosomal protein S6
+Macromolecule #37: 30S ribosomal protein S7
+Macromolecule #38: 30S ribosomal protein S8
+Macromolecule #39: 30S ribosomal protein S9
+Macromolecule #40: 30S ribosomal protein S10
+Macromolecule #41: 30S ribosomal protein S11
+Macromolecule #42: 30S ribosomal protein S12
+Macromolecule #43: 30S ribosomal protein S13
+Macromolecule #44: 30S ribosomal protein S14
+Macromolecule #45: 30S ribosomal protein S15
+Macromolecule #46: 30S ribosomal protein S16
+Macromolecule #47: 30S ribosomal protein S17
+Macromolecule #48: 30S ribosomal protein S18
+Macromolecule #49: 30S ribosomal protein S19
+Macromolecule #50: 30S ribosomal protein S20
+Macromolecule #52: TELITHROMYCIN
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 1.425 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)